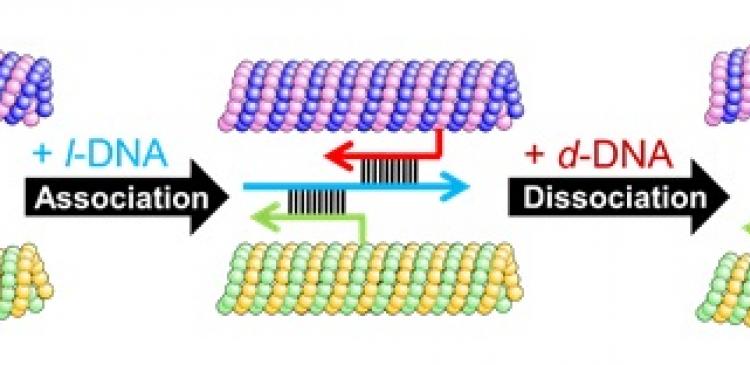
Microtubules (MT) interact with each other through DNA molecules while moving around on the kinesin motors. (Keya J. J. et al., Nature Communications, January 31, 2018)
[Joint press release by Hokkaido University and Kansai University]
The world’s smallest “swarm robot” measures 25 nanometers in diameter and 5 micrometers in length, and exhibits swarming behavior resembling motile organisms such as fish, ants and birds.
“Swarm robots are one of the most elusive subjects in robotics,” says Akira Kakugo of the research team at Hokkaido University. “Fish schools, ant colonies and bird flocks show fascinating features that cannot be achieved by individuals acting alone. These include the formation of complex structures, distinct divisions of labor, robustness and flexibility, all of which emerge through local interactions among the individuals without the presence of a leader.” Inspired by these characteristics, researchers have been working to develop micro-scale swarm robots.
In the present study, Kakugo and his collaborators have built a molecular system that is composed of the three essential components of a robot: sensors, information processors and actuators. They used cellular proteins called microtubules and kinesins as the actuator, and DNA as the information processor. Microtubules are filamentous proteins that serve as the railways in the cellular transportation system, while kinesins are motor proteins that run on the railways by consuming chemical energy obtained from hydrolysis of adenosine triphosphate (ATP). The team took a reverse strategy and built a system in which microtubules move randomly on a kinesin coated surface.
A major challenge in swarm robotics is the construction of a large number of individual robots capable of programmable self-assembly. The team addressed this issue by introducing DNA molecules into the system that are known to hybridize when they have a complementary sequence. The chemically synthesized DNA molecules with certain programs in their sequences are conjugated to the microtubules labeled with green or red fluorescence dye.
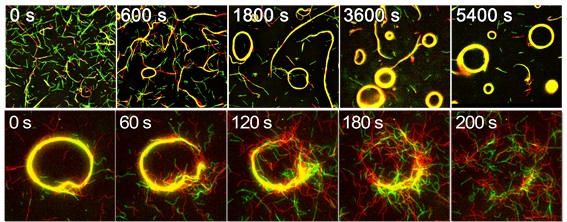
The team then monitored the motions of the DNA-conjugated microtubules gliding on a kinesin coated surface. Initially, five million microtubules moved without any interactions with each other. They then added single-strand linker DNA (l-DNA), programmed to initiate interactions among the DNA-attached microtubules. Upon introduction of the l-DNA, the microtubules began to assemble and formed swarms of a much larger size than the microtubules. When another single-strand DNA (d-DNA), programmed to dissociate the swarms was added, the microtubule swarms disappeared soon. This demonstrated that swarming of a large number of microtubules can be reversibly regulated by selectively providing the input DNA signal in the system.
Moreover, they added a photosensitive sensor to the system, azobenzene attached to the DNA molecules. They utilized isomerization of the azobenzene that occurs reversibly in response to irradiation of visible or ultraviolet light to switch on or off the interaction between DNA molecules. This enabled the photo-irradiation induced switching between the solitary and swarm state of the microtubules. The team also demonstrated that the swarms of microtubules move with a translational or rotational motion depending on the rigidity of the microtubules.
“This is the first evidence showing that swarming behavior of molecular robots can be programmed by DNA computing. The system acts as a basic computer by executing simple mathematical operations, such as AND or OR operations, leading to various structures and complex motions. It is expected that such a system contributes in developing artificial muscles and gene diagnoses, as well as building nano-machines in the future,” Kakugo commented.
Contacts:
Associate Professor Akira Kakugo
Department of Chemistry, Hokkaido University
Department of Biomedical Engineering, Columbia University
Email: kakugo[at]sci.hokudai.ac.jp
Associate Professor Akinori Kuzuya
Department of Chemistry and Materials Engineering
Kansai University
Email: kuzuya[at]kansai-u.ac.jp
Naoki Namba (Media Officer)
Global Relations Office
Institute for International Collaboration
Hokkaido University
Tel: +81-11-706-2185
Email: pr[at]oia.hokudai.ac.jp
Kansai University Public Affairs Division
Email: kouhou[at]ml.kandai.jp
URL: http://www.kansai-u.ac.jp/English/index.html
The images show the flexible microtubules forming swarms with circular motions (top), and them dissociating in response to the d-DNA input signal (bottom). The movie (linked below) shows the motility, swarming and dissociation of the flexible (low rigidity) microtubules. (Keya J. J. et al., Nature Communications, January 31, 2018)