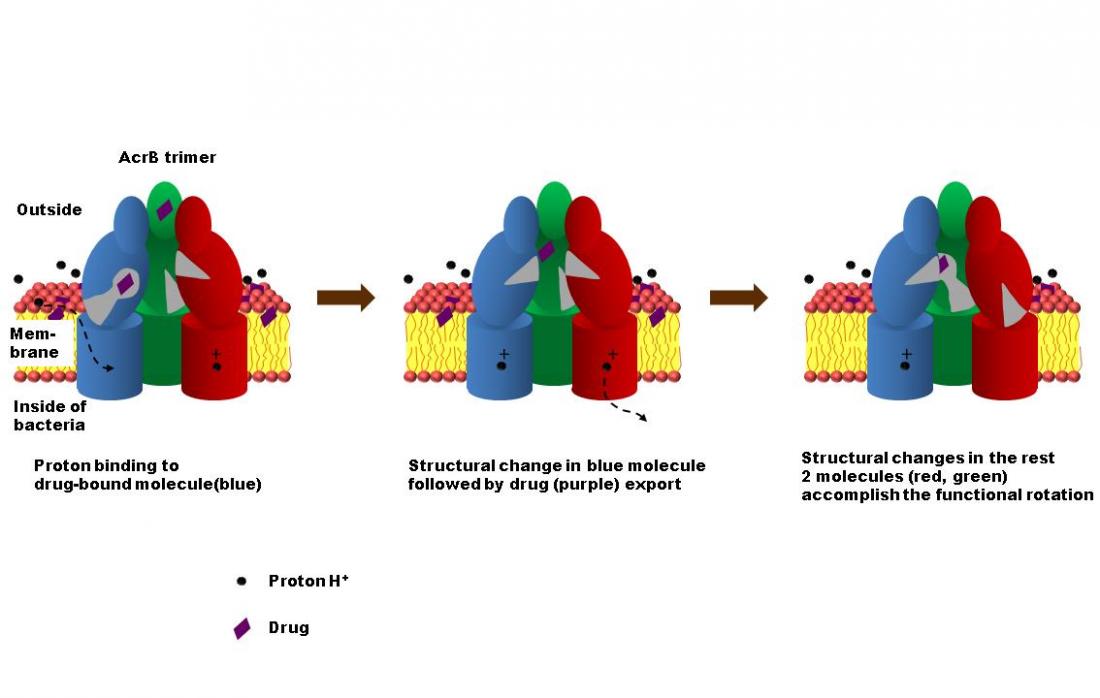
Figure 1: Triggering of AcrB drug export and functional rotation by proton-binding.
Molecular simulations confirm role of functional rotation in multidrug resistance
A new molecular simulation technique developed by researchers at RIKEN and Kyoto University has confirmed for the first time the function of the transporter protein AcrB in E. coli multidrug resistance. The result marks a key step in efforts to combat antibiotic resistance, demonstrating the power of computer simulation as an essential tool for basic research.
The capacity of certain bacteria to resist a wide variety of drugs and chemicals, known as “multidrug resistance”, has attracted widespread attention for the threat it poses to global health. One of the key factors that cause such resistance is the overexpression of efflux pumps, protein transporters in the cytoplasmic membrane that flush toxic substances out of the cell.
To clarify the role of efflux pumps in multidrug resistance, the researchers modeled the dynamics of AcrB, a transporter in E. coli which enables the bacteria to expel a wide range of target substances, including many antibiotics. Earlier research characterized AcrB as a prism-shaped homo-trimer of three protein subunits with conformations corresponding to three states in a transport cycle governing access to, binding and extrusion of target substances from the cell. Evidence for this so-called “functional rotation” hypothesis, however, was lacking.
Using a new coarse-grain molecular simulation technique, the research group has now produced this crucial evidence for the first time, and has pinpointed its driving force in proton-binding to AcrB’s drug-bound molecule. The findings also reconcile previous results, which in 2002 described AcrB as a symmetric structure and then in 2006 an asymmetric one, by suggesting that each study describes a different state of the transporter, whose conformation changes during drug dissociation.
Reported in the journal Nature Communications, this verification of the functional rotation mechanism marks a key step forward toward tackling key problems of multidrug resistance. It also hints at breakthroughs to come with the launch of RIKEN’s new Next-Generation Supercomputer (“K Computer”), set to be unveiled in 2012.
For more information, please contact:
Dr. Ryutaro Himeno
Integrated Simulation of Living Matter Group
RIKEN Computational Science Research Program
Tel: +81-(0)48-467-9321 / Fax: +81-(0)48-462-4634
Dr. Shoji Takada
Department of Biophysics, Graduate School of Science
Kyoto University
Tel: +81-(0)75-753-4220 / Fax: +81-(0)75-753-4222
Ms. Tomoko Ikawa (PI officer)
Global Relations Office
RIKEN
Tel: +81-(0)48-462-1225 / Fax: +81-(0)48-463-3687
Email: [email protected]
Reference:
Xin-Qiu Yao, Hiroo Kenzaki, Satoshi Murakami, and Shoji Takada. Drug export and allosteric coupling in a multidrug transporter revealed by molecular simulations. Nature Communications 1: 117 (2010)(8 pages). doi: 10.1038/ncomms1116 (2010).