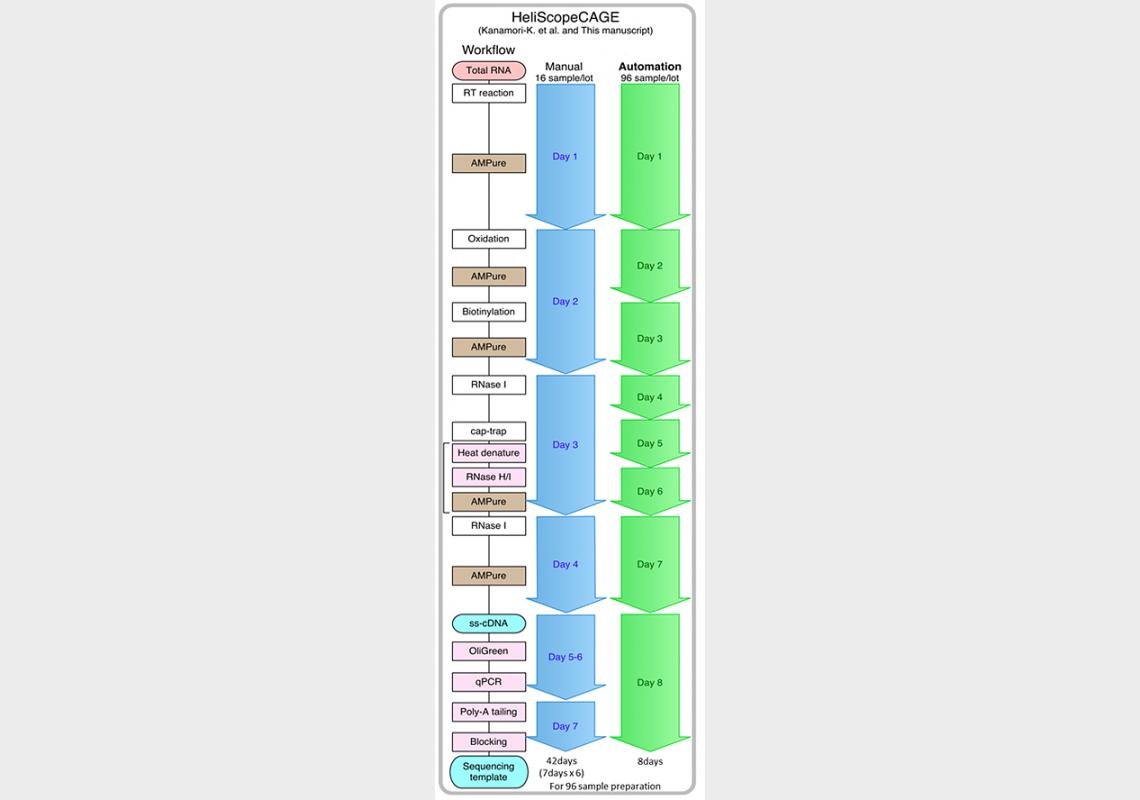
Figure 1 - Comparison of time frame between manual and automated processes
The workflow uses Cap Analysis of Gene Expression (CAGE), a unique method developed at the OSC for determining transcriptional starting sites in the genome and their expression levels.
A key obstacle in DNA sequencing is the long time required for sample preparation, especially when compared with how fast next-generation sequencers are able to sequence DNA. Now, researchers at the OSC, who previously adapted the CAGE method to the HeliScope single molecule sequencer, have reduced that time using a new automated sample preparation system. Using the new system, sample preparation of 96 CAGE cDNA libraries, which would normally take a manual operator 42 days, is done in only 8 days.
The researchers further demonstrated, through a comparison of results from the analysis of transcriptional starting sites and their expression levels using CAGE on the HeliScope single molecule sequencer, that results using the new preparation system are reproducible and comparable to results using manual preparation. The new automated cDNA preparation system can also be applied to a variety of other methods and sequencers, including CAGE on the Illumina/SOLD platform, RNA-seq and full-length cDNA generation.
----------------------------------------------------------------
For more information, please contact
Masayoshi Itoh
LSA Technology Development Group
RIKEN Omics Science Center (OSC)
Tel: +81-(0)45-503-9222 / Fax: +81-(0)45-503-9216
Yokohama Planning Section
RIKEN Yokohama Research Promotion Division
Tel: +81-(0)45-503-9117 / Fax: +81-(0)45-503-9113
Global Relations Office
RIKEN
Tel: +81-(0)48-462-1225 / Fax: +81-(0)48-463-3687
Mail: [email protected]
----------------------------------------------------------------------
Reference:
Masayoshi Itoh, Miki Kojima, Sayaka Nagao-Sato, Eri Saijo, Timo Lassmann, Mutsumi Kanamori-Katayama, Ai Kaiho, Marina Lizio, Hideya Kawaji, Piero Carninci, Alistair R R Forrest, Yoshihide Hayashizaki. "Automated workflow for preparation of cDNA for cap analysis of gene expression on a single molecule sequencer." PLoS ONE, 2012, DOI: 10.1371/journal.pone.0030809
----------------------------------------------------------------------
About RIKEN
RIKEN is Japan's flagship research institute devoted to basic and applied research. Over 2500 papers by RIKEN researchers are published every year in reputable scientific and technical journals, covering topics ranging across a broad spectrum of disciplines including physics, chemistry, biology, medical science and engineering. RIKEN's advanced research environment and strong emphasis on interdisciplinary collaboration has earned itself an unparalleled reputation for scientific excellence in Japan and around the world.
About the RIKEN Omics Science Center
Omics is the comprehensive study of molecules in living organisms. The complete sequencing of genomes (the complete set of genes in an organism) has enabled rapid developments in the collection and analysis of various types of comprehensive molecular data such as transcriptomes (the complete set of gene expression data) and proteomes (the complete set of intracellular proteins). Fundamental omics research aims to link these omics data to molecular networks and pathways in order to advance the understanding of biological phenomena as systems at the molecular level.
Here at the RIKEN Omics Science Center, we are developing a versatile analysis system, called the "Life Science Accelerator (LSA)", with the objective of advancing omics research. LSA is a multi-purpose, large-scale analysis system that rapidly analyzes molecular networks. It collects various genome-wide data at high throughput from cells and other biological materials, comprehensively analyzes experimental data, and thereby aims to elucidate the molecular networks of the sample. The term "accelerator" was chosen to emphasize the strong supporting role that this system will play in supporting and accelerating life science research worldwide.
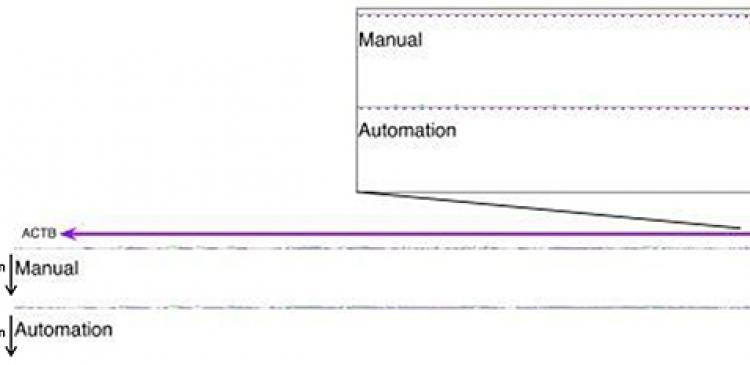
Figure 2 - Genomic view of a selected gene for comparison between manual and automated methods CAGE mapped read counts were displayed as linear scale histogram on ACTB gene. The transcription initiation regions are magnified and demonstrate that the distribution of transcription starting sites and their expression levels are consistent between manual and automated methods.