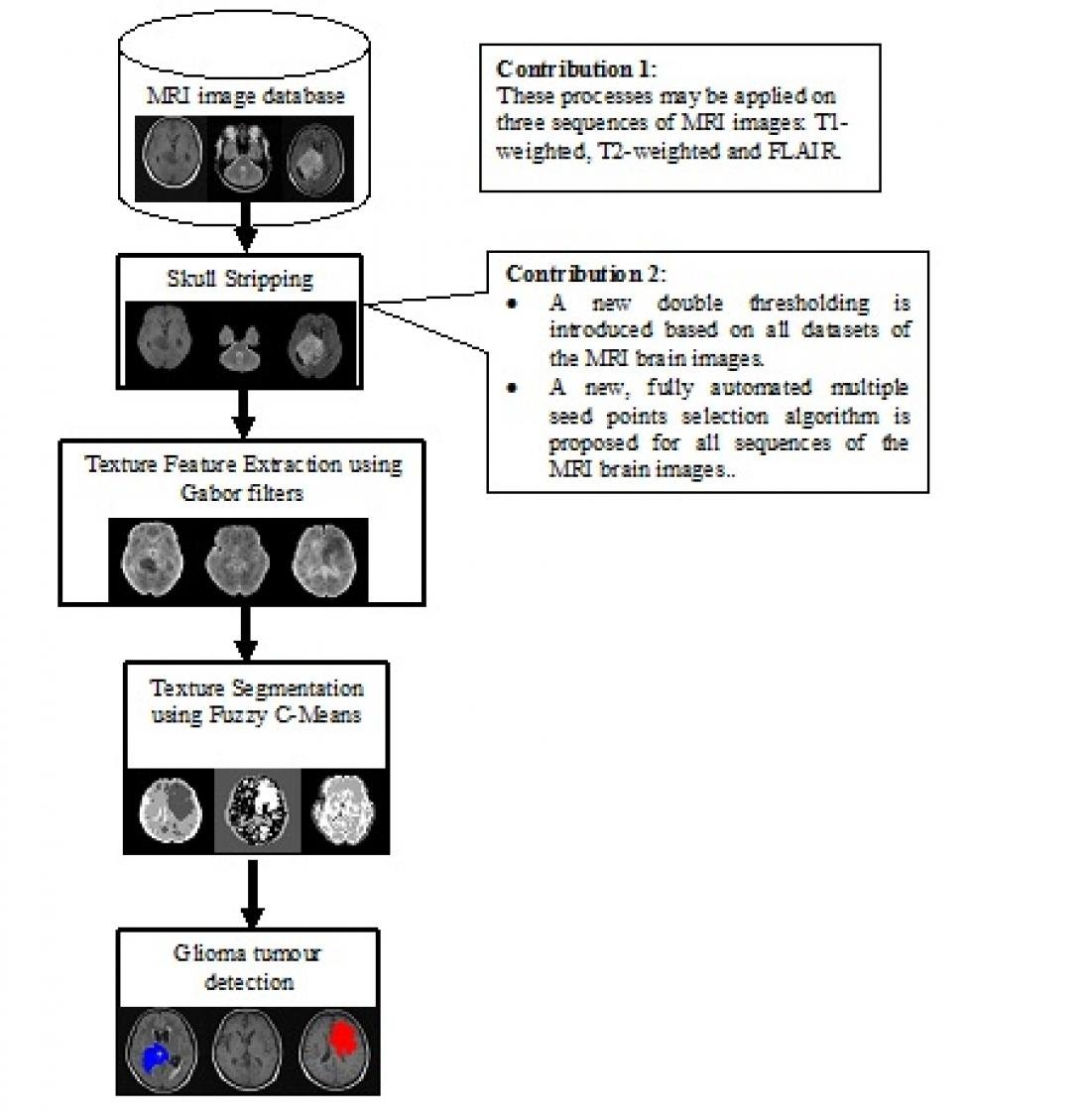
Figure 1: Process flow of the research
Recent statistics showed that five percent of Malaysians have been diagnosed with brain tumours with glioma being the most common type. Radiologists commonly use MRI image sequences to detect glioma clinically by examining the abnormalities on T1-Weighted, T2-Weighted and Fluid Attenuated Inversion Recovery (FLAIR) images. However, when the tumour cannot be detected visually, they will inject a contrast agent of gadolinium to enhance the image modality. This process delays acquisition of results is at a higher cost and imposes side effects to the patients. Therefore, this study proposes a non-invasive solution by utilizing spectral texture features of the MRI images in detecting the tumour in all three sequences of T1, T2-Weighted and FLAIR images.
There are four phases involved in this project which are data collection, skull-stripping, texture feature extraction, and segmentation. For data collection, a total of 126 MRI images of adults ranging from 18 to 60 years old are obtained from Hospital Sungai Buloh and Hospital Tengku Ampuan Rahimah Klang in Selangor. Ninety MRI image sequences of T1-Weighted, T2-Weighted and FLAIR are used for skull-stripping experiments and results showed that mathematical morphology method outperformed region growing at an accuracy rate of 96%.
A new double thresholding algorithm and a fully automated multiple seed points selection algorithm that works on all three MRI image sequences were proposed. For texture feature extraction, we tested three features that are inverse Fast Fourier Transform (IFFT), texture energy and transformed IFFT. Experiments conducted on 64 MRI images of all sequences showed that texture energy is the best texture feature to be used in glioma segmentation Fuzzy C-Means clustering algorithm is then used to segment texture energy features from 126 MRI brain images of all sequences.
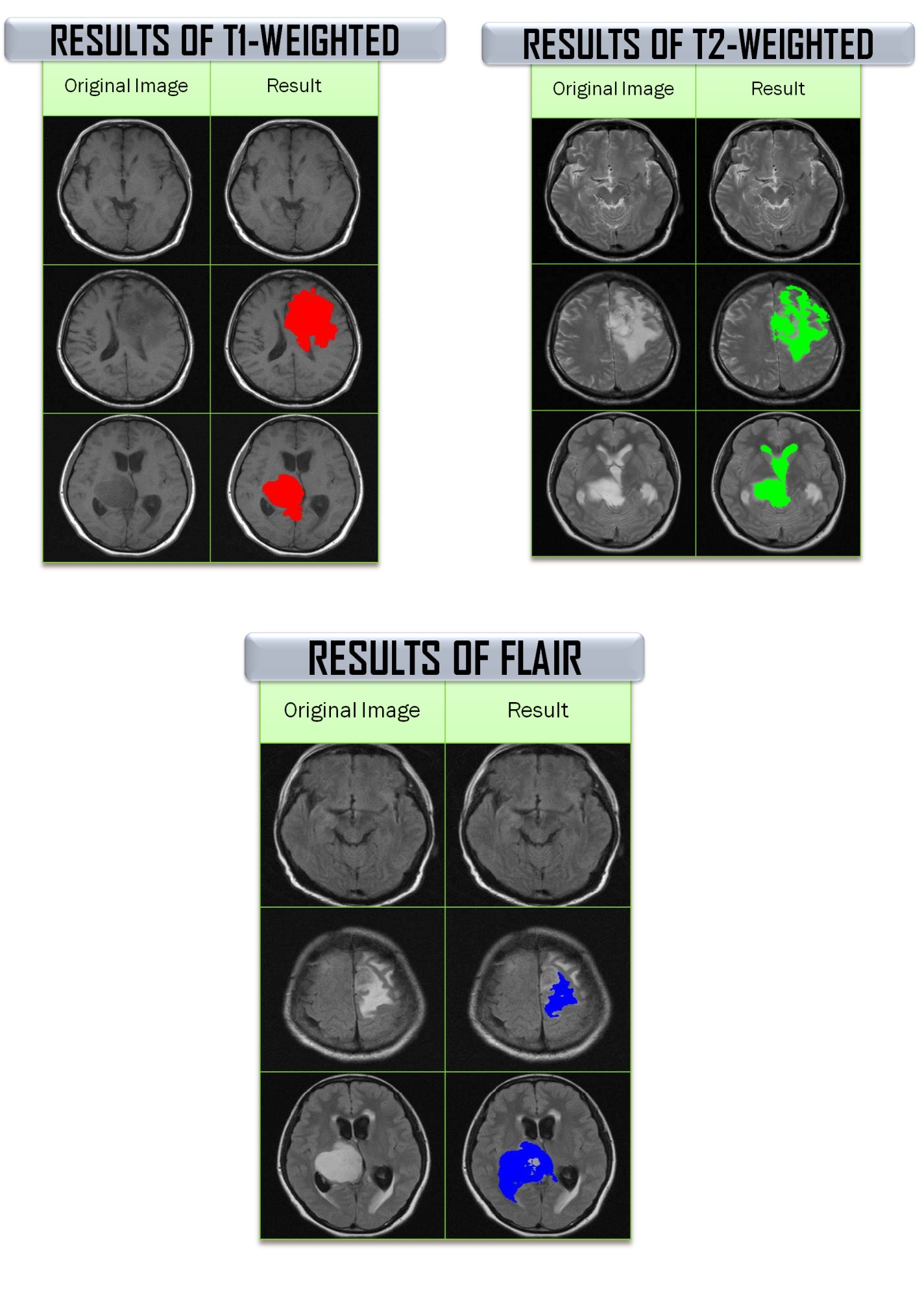
Results are then evaluated by an expert radiologist and it showed that the glioma brain tumour is detected. Therefore, Fuzzy C-Means clustering method has been identified as promising methods of glioma brain tumour detection at an accuracy rate of 76%, 86% and 79% for T1-Weighted, T2-Weighted and FLAIR images, respectively.
Other than glioma tumour detection, the proposed methods may be used for other types of brain abnormalities detection on MRI images. Unlike other methods that normally utilized one MRI sequence, our proposed methods worked on all three sequences of MRI images. A prototype of the automated glioma brain tumour detection was developed and is ready to be installed and used by radiologist, neurologist, medical researchers and students.
Nursuriati Jamil, Rosniza Roslan
Faculty of Computer and Mathematical Sciences, MARA University of Technology,
Shah Alam, Selangor.
[email protected]