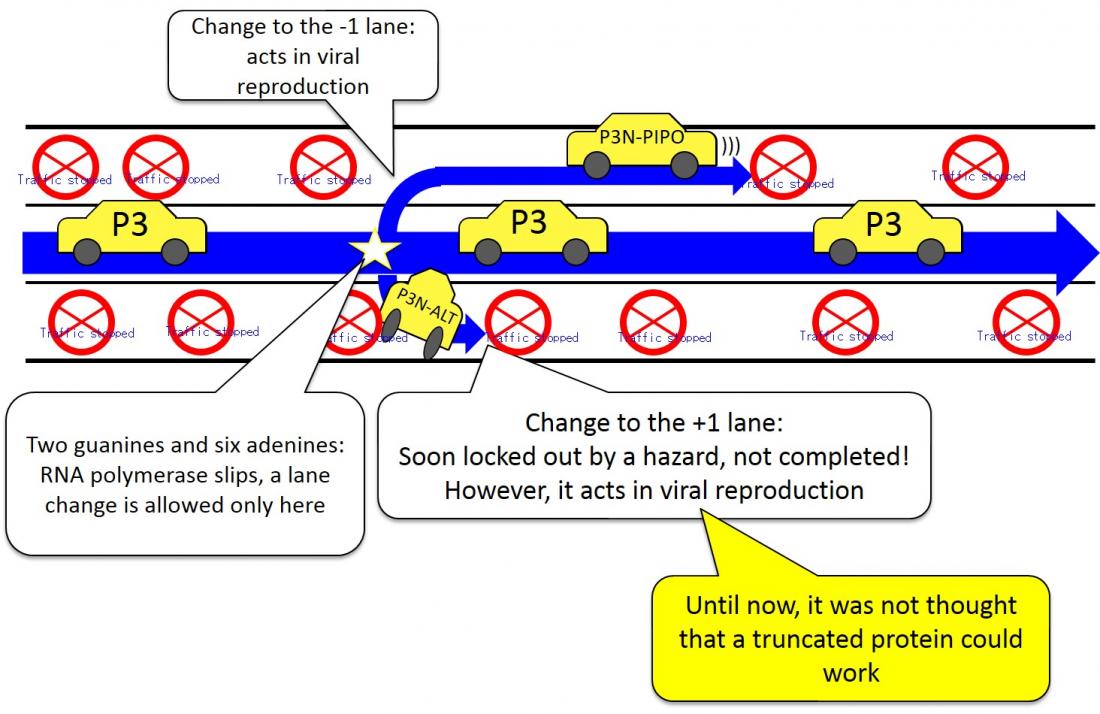
Image of how the truncated gene P3N-ALT is formed: Even though it soon stops (the translation into a protein is ended), there is a car that changes lanes to the +1 lane (reading frame) at a base sequence that contains two guanines and six adenines (G2A6) that occurs in the middle of the center line (the large ORF in the viral genome), and a small amount of P3N-ALT protein is produced. This truncated gene P3N-ALT turns out to be necessary for viral infection and reproduction.
Key Points:
- Clover yellow vein virus produces a truncated form of the P3 protein that is indispensable for viral infection and multiplication.
- The truncated protein is produced via an unusual slippage of RNA polymerase at the G2A6 sequence, causing a shift in the genetic code-reading frame from the normal P3, to one that had not been thought to produce protein.
- A diverse range of RNA viruses possess the G2A6 or similar A-stretch sequences, suggesting that they also utilize a similar strategy during their infection.
Background:
Viruses that have RNA (ribonucleic acid) as their genome must pack the genes necessary for viral infection and reproduction into small genomes.
They use a variety of strategies to express them. When the clover yellow vein virus creates a copy of itself, it deletes one base from a sequence of two guanines and six adenines (G2A6) within the gene P3.
As a result, a truncated P3 gene (named P3N-ALT) is expressed, in which the portion of P3 downstream of the G2A6 is lost. Also, P3N-ALT is required for viral infection and reproduction.
These facts have been elucidated at this time by the research group composed of Researcher Yuka Komoda (Hagiwara), Instructor Kenji Nakahara, and Professor Satoshi Naito of Hokkaido University, and Specially-Appointed Assistant Professor Masanao Sato et al. of Keio University.
Many similar sequences for which a base is possible to be inserted or deleted have been found in the genomes of other plant and animal RNA viruses, so it is thought that truncated genes that are necessary for infection and reproduction may be expressed in other viruses as well.
Anticipated Outcomes:
Until now, it was not expected that a truncated gene like P3N-ALT would serve a function required for viral infection and reproduction. This research has demonstrated for the first time that the truncated gene P3N-ALT does work for viral infection and reproduction.
It is possible that many other RNA viruses also express truncated genes, and by considering this possibility in the research, we can expect new discoveries related to viral infection and reproduction, as well as pathogenicity.
Inquiries:
Lecturer Kenji NAKAHARA
Laboratory of Pathogen-Plant Interactions, Research Group of Plant Breeding Science, Division of Fundamental AgriScience Research, Research Faculty of Agriculture, Hokkaido University
knakahar[at]res.agr.hokudai.ac.jp
Publications:
Truncated yet functional viral protein produced via RNA polymerase slippage implies underestimated coding capacity of RNA viruses, Scientific Reports (02.22.2015)