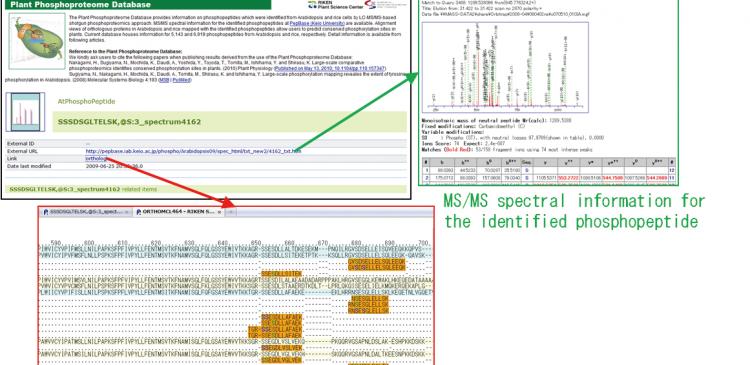
Figure 1: Overlap between rice and Arabidopsis phosphoproteomes
Researchers at RIKEN and Keio University have shown that even the most widely-varying species
of plants share remarkable similarities in the composition of proteins in them that undergo
phosphorylation, a regulatory mechanism involved in various cellular phenomena. A database
released by the group, with information on over three thousand phosphorylated proteins and
phosphorylation sites in rice, opens new doors in the study and engineering of plants.
The addition of a phosphate group to a protein, known as phosphorylation, plays a vital role in
regulating cellular phenomena and as a mediator of signaling pathways in the cell. The function of
this process in regulating plant growth and development in particular makes it highly attractive for
plant engineering, yet existing resources on phosphorylation are limited to model plants such as
Arabidopsis, beyond which their applicability is unclear.
To expand the range of uses for these resources, the research group set out to determine the degree
to which phosphorylation mechanisms are conserved across two very different plant species:
Arabidopsis, from the family of flowering plants known as dicotyledons (dicots), and rice, from
the family known as monocotyledons (monocots). Their large-scale analysis on rice, the first ever,
identified a total of 3393 different types of proteins regulated by phosphorylation and their
phosphorylation sites, of which more than half, they showed, are shared by Arabidopsis.
The surprising discovery that these two very different plants exhibit significant similarities in their
mechanisms of phosphorylation suggests that information on the “phosphoproteome” of one
species can be applied to others, greatly contributing to applications in plant engineering.
Data leading to the discovery has been made available to the public in an open-access database,
the Plant Phosphoproteome Database, released online on May 12.
For more information, please contact:
Dr. Ken Shirasu
Plant Immunity Research Group
RIKEN Plant Science Center (PSC)
Tel: +81-(0)45-503-9574 / Fax: +81-(0)45-503-9573
Dr. Hirofumi Nakagami
Plant Proteomics Research Unit
RIKEN Plant Science Center (PSC)
Tel: +81-(0)45-503-9424 / Fax: +81-(0)45-503-9573
Ms. Tomoko Ikawa (PI officer)
Global Relations Office
RIKEN
Tel: +81-(0)48-462-1225 / Fax: +81-(0)48-462-4715
Email: [email protected]
About the RIKEN Plant Science Center
With rapid industrialization and a world population set to top 9 billion within the next 30 years,
the need to increase our food production capacity is more urgent today than it ever has been before.
Avoiding a global crisis demands rapid advances in plant science research to boost crop yields and
ensure a reliable supply of food, energy and plant-based materials.
The RIKEN Plant Science Center (PSC), located at the RIKEN Yokohama Research Institute in
Yokohama City, Japan, is at the forefront of research efforts to uncover mechanisms underlying
plant metabolism, morphology and development, and apply these findings to improving plant
production. With laboratories ranging in subject area from metabolomics, to functional genomics,
to plant regulation and productivity, to plant evolution and adaptation, the PSC’s broad scope
grants it a unique position in the network of modern plant science research. In cooperation with
universities, research institutes and industry, the PSC is working to ensure a stable supply of food,
materials, and energy to support a growing world population and its pressing health and
environmental needs.