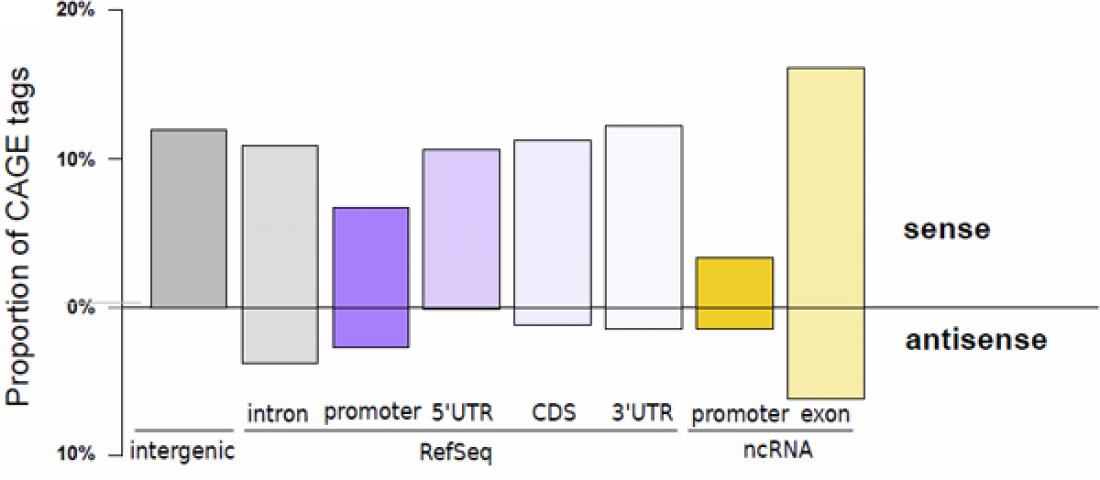
Figure 1 MOE transcription start sites recapitulate known transcript initiation and reveal the extent of non-coding transcripts.
Histogram depicting the proportion of tags aligned to the proximal promoter of transcript models (defined as the region spanning from the 5' end to 500 bp upstream), the 5' UTR, the coding sequence (CDS), the 3' UTR (in decreasing purple colors), the proximal promoter of FANTOM3 non-coding RNA (in orange), and the FANTOM3 non-coding RNA (in light orange). The upper part of the bar plot shows TSSs located on the same strand as the annotation, while the lower part depicts TSSs located on the opposite strand. The percentage of TSSs that do not colocalize with any of those annotations is represented by the grey bar.
In mouse, odor is sensed by the main olfactory epithelium (MOE) by about 1100 types of olfactory receptors that are expressed by olfactory sensory neurons. Interestingly, each sensory neuron expresses only a single type of olfactory receptors, whose selective expression mechanism remains largely unknown. The population of olfactory sensory neurons that express a given olfactory receptor is small, which makes transcription analysis difficult.
Researchers at the RIKEN Omics Science Center recently developed nanoCAGE (CAGE: Cap Analysis of Gene Expression), the only technology that can comprehensively identify precise TSSs of both protein-coding and non-coding capped mRNAs and quantify their individual levels of expression starting from tiny biological samples of only a few nanograms of RNA (Plessy et al., Nature Methods, 7, 528-534, 2010). By using nanoCAGE on the MOE, the researchers succeeded in identifying 87.5% of the olfactory receptor gene TSSs. The results show for the first time that olfactory receptor genes contain hundreds of non-coding RNAs, suggesting that these RNAs may play important roles in the transcriptional regulation of olfactory receptors.
Dr. Piero Carninci commented, "Combined with CAGE, nanoCAGE technology provides a new opportunity to unveil gene networks in the nervous system using omics approaches."
The research is published in the journal Genome Research.
--------------------------------------------------------------------
REFERENCE
Plessy C, Pascarella G, Bertin N, Akalin A, Carrieri C, Vassalli A, Lazarevic D, Severin J, Vlachouli C, Simone R, Faulkner GJ, Kawai J, Daub CO, Succhelli S, Hayashizaki Y, Mombaerts P, Lenhard B, Gustincich S, Carninci P. "Promoter architecture of mouse olfactory receptor genes." Genome Research, 2011, DOI: 10.1101/gr.126201.111
---------------------------------------------------------------------
CONTACT
Piero Carninci
Functional Genomics Technology Team
RIKEN Omics Science Center
Tel: +81-(0)45-503-9222 / Fax: +81-(0)45-503-9216
Mail
Yokohama Planning Section
RIKEN Yokohama Research Promotion Division
Tel: +81-(0)45-503-9117 / Fax: +81-(0)45-503-9113
Global Relations Office
RIKEN
Tel: +81-(0)48-462-1225 / Fax: +81-(0)48-463-3687
Mail: [email protected]