A joint research team, affiliated with UNIST has identified a new metabolic pathway, in which microorganisms utilize single carbon (C1) gases (CO and CO2) as a feedstock. The new metabolic pathway is thought to be the most energetically efficient pathway, compared to the existing ones, and thus is expected to be used in a variety of industrial applications that involved the conversion of C1 gas into value-added biochemicals.
Published in the March 13th issue of PNAS (Proceedings of the National Academy of Sciences of the United States of America), this research has been jointly carried out by Professor Donghyuk Kim (School of Energy and Chemical Engineering, UNIST) and Professor Byung-Kwan Cho (Department of Biological Sciences, KAIST), and thus participated by Dr. Yoseb Song (Department of Biological Sciences, KAIST), as the first author.
There are currently six autotrophic CO2 fixation pathways, capable of converting C1 gas into organic compounds and one representative example is the photosynthesis in plants. Among those CO2-fixing metabolic pathways in nature, the linear wood-ljungdahl pathway (WLP) in phylogenetically diverse acetateforming acetogens is known to be the most energetically efficient pathway to fix C1 compounds. In particular, acetogens play an important role in the global carbon cycle, with nearly 1013 kg (100 billion US tons) of acetic acid being formed annually.
However, the growth rate of acetogens is 10 times slower than that of industrial microorganisms, such as E. coli. And this puts a limit on its use as industrial microorganisms for the conversion of C1 gas into useful biochemical products. Accordingly, many studies on a new and more effective CO2 fixation have been carried out.
The research team paid special attention to the growth rate of Clostridium drakei, which was faster than that of the other microorganisms, when accompanied by CO2 absorption. And, through this, they expected they might find clues to enhance the C1 gas conversion efficiency.
Figure 1. Acetyl-CoA production ratio in wild-type and knockout strains.
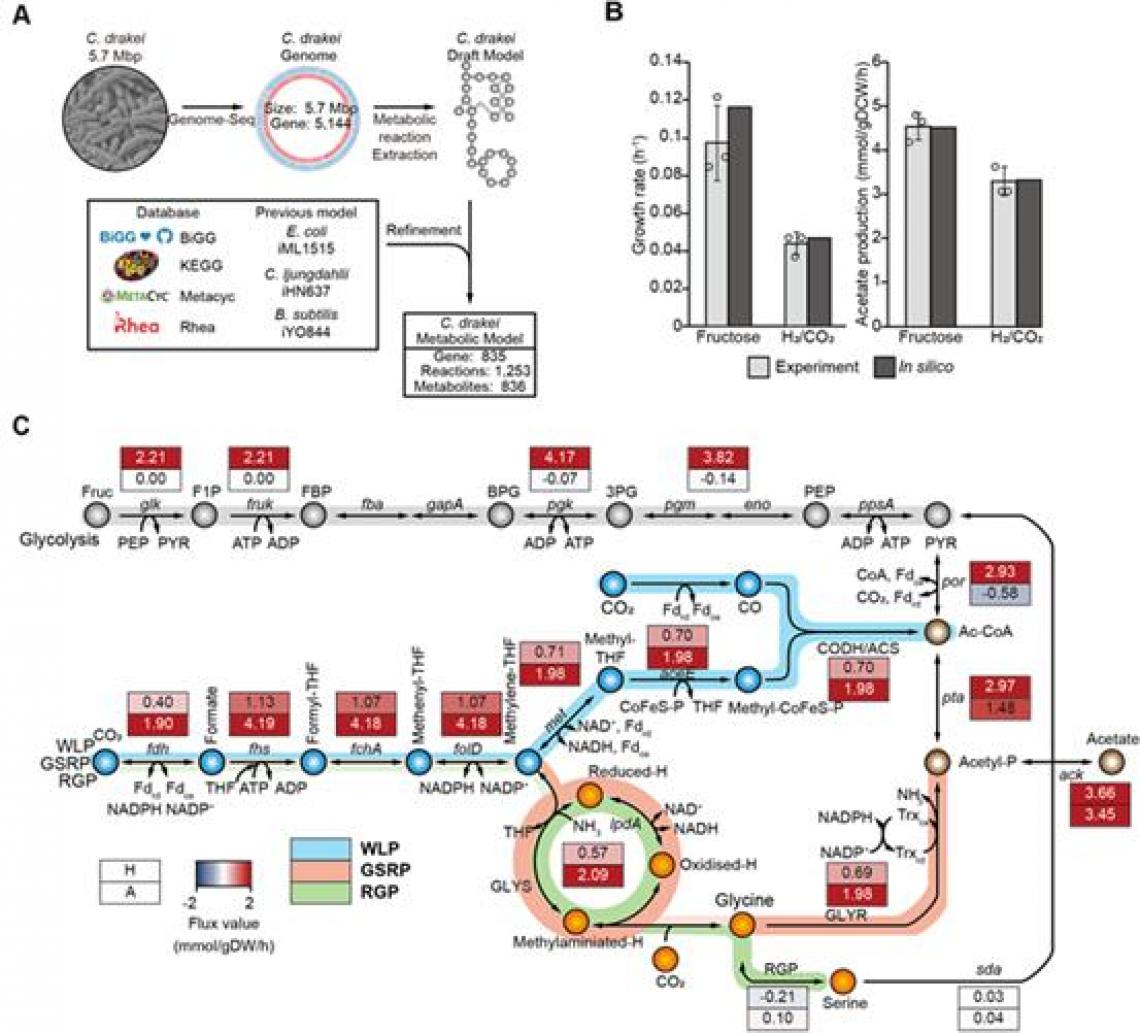
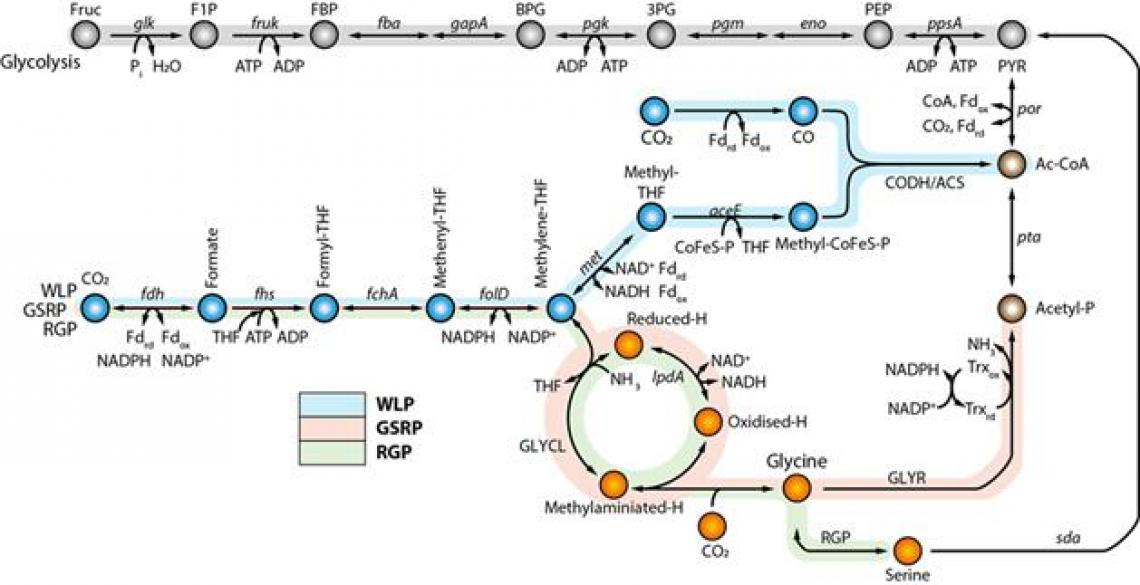
In this study, using the reconstructed genome-scale metabolic model iSL771 based on the completed genome sequence, transcriptomics, 13C isotope-based metabolite-tracing experiments, biochemical assays, and heterologous expression of the pathway in another acetogen, the research team discovered that the WLP and the glycine synthase pathway are functionally interconnected to fix CO2, subsequently converting CO2 into acetyl-CoA, acetyl-phosphate, and serine.
Moreover, the functional cooperation of the pathways enhances CO2 consumption and cellular growth rates via bypassing reducing power required reactions for cellular metabolism during autotrophic growth of acetogens.
Figure 2. Construction of the genome-scale metabolic network model of C. drakei (iSL771).
“With the new CO2-fixing metabolic pathway, we shall overcome limitations in the biosynthesis for the production of high value-added compounds, brought by the slow growth rate of acetogens,” says Professor Kim.
This work has been supported by the Intelligent Synthetic Biology Centre of the Global Frontier Project and the C1 Gas Refinery Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Science and ICT (MSIT) and KAIST Cross-Generation Collaborative Research Group (KAIST Biodesign Engineering Laboratory).