Tokyo, Japan – You may be familiar with the art of origami, in which paper is intricately folded to create shapes. But did you know that proteins in the human body also undergo an intricate folding process that is essential to their structure and function? Recently, researchers in the U.S. and Japan have shed new light on protein folding stability, or the propensity of a protein to maintain its folded shape – a factor at the center of diseases such as cancer, Alzheimer’s disease, and cystic fibrosis.
In a new study published in Nature, a research team from Northwestern University and recently joining the Institute for Industrial Science, The University of Tokyo have developed a new high-throughput approach known as cDNA display proteolysis to evaluate the folding stability of nearly a million proteins in a single experiment.
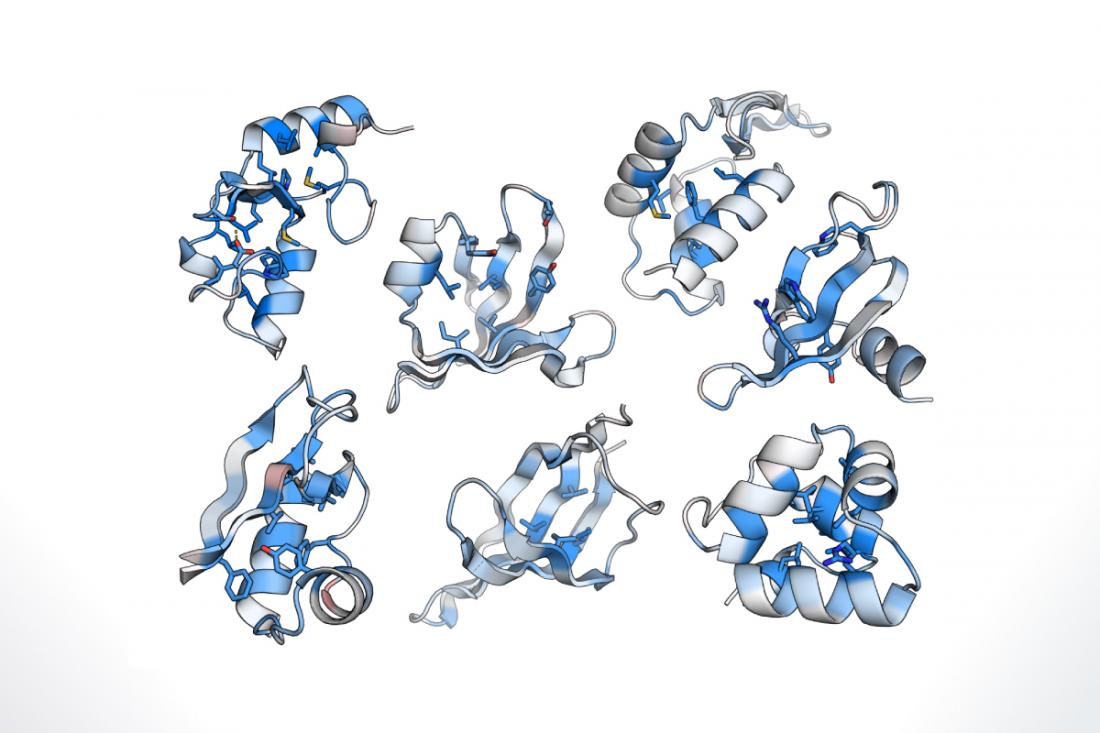
A protein is initially generated as a single chain of amino acids that is then folded into a three-dimensional shape. Failure to fold properly or maintain this three-dimensional structure can disrupt protein function and lead to disease. Insight into how protein folding stability is maintained will therefore shed new light on diseases involving misfolded proteins. However, it has previously been difficult to evaluate protein folding stability in an efficient and large-scale manner. Therefore, the research team sought to develop a platform to assess protein folding stability in a reproducible, high-throughput way.
“We began with a technique in which proteins are attached to their own DNA,” says lead author of the study Kotaro Tsuboyama. “Using DNA libraries, we generated a large number of these protein–DNA complexes and treated them with enzymes that destroy unfolded proteins. The intact proteins, which were able to maintain their folded structures during enzyme treatment, were then identified using DNA sequencing.”
This method allowed the research team to evaluate the stability of up to 900,000 protein sequences in a single test tube. To examine how individual elements within a protein sequence affect folding stability, the researchers used this method to analyze a series of natural and designed protein domains.
“We were able to identify a number of factors that contribute to protein stability,” says senior author Gabriel J. Rocklin at Northwestern University. “We also used our approach to analyze the effects of specific mutations in protein sequences, and to identify determinants of stability in designed proteins, providing insight that can help advance protein design methods in the future.”
While previous methods for assessing protein stability have been limited to evaluating single-protein sequences, the cDNA display proteolysis method permits the evaluation of many proteins in a single experiment, supplying an unprecedented amount of information regarding protein stability. This approach may advance the development of new predictive models of protein folding, which may further our understanding of diseases involving protein misfolding.
###
The article, “Mega-scale experimental analysis of protein folding stability in biology and design,” was published in Nature at DOI: 10.1038/s41586-023-06328-6.
About Institute of Industrial Sciene, The University of Tokyo
The Institute of Industrial Science, The University of Tokyo (UTokyo-IIS) is one of the largest university-attached research institutes in Japan. UTokyo-IIS is comprised of over 120 research laboratories—each headed by a faculty member—and has over 1,200 members (approximately 400 staff and 800 students) actively engaged in education and research. Its activities cover almost all areas of engineering. Since its foundation in 1949, UTokyo-IIS has worked to bridge the huge gaps that exist between academic disciplines and real-world applications.