Bifidobacteria and pathogenic Escherichia coli O157. Bifidobacteria consume polysaccharides (dietary fibers) and produce acetate. The resulting acetate protects the mucosal epithelium of the large intestine to enhance its defense against infection. As a result, epithelial cell death caused by O157 is prevented, decreasing the risk of O157-produced shiga toxin entering the body by bypassing the bowel mucosal epithelium. Meanwhile, O157 consumes the aspartic acid and serine produced by bifidobacteria to produce succinate and acetate. Bifidobacteria and O157 have a weakly symbiotic relationship.
Heading the Advanced NMR Metabomics Research Team in the Metabolomics Research Division at the RIKEN Plant Science Center, Jun Kikuchi is engaged in analyzing metabolism within living organisms, mainly using nuclear magnetic resonance (NMR) spectroscopy. He uses a wide range of samples in his research, including not only higher plants, but also algae, complex microbial systems, insects, mice, fish, foods and beverages. “The goal of my research is to gain a comprehensive understanding of metabolic dynamics in a wide variety of organisms, with a view to elucidating the universal laws of nature,” says Kikuchi. “This will lead to green innovations that may increase the potential of organisms to metabolize valuable substances, and to innovations that will contribute to better health. I want to be an innovator in these areas.” Kikuchi is engaged in the technical development of NMR analysis, specifically for metabolic dynamics.
Genomics, proteomics and now metabolomics
Twenty-seven instruments are available at the nuclear magnetic resonance facility of the RIKEN Yokohama Institute, making it the foremost facility of its kind in the world. “Many of the NMR systems here are used to analyze protein structure,” says Kikuchi. “As a researcher at the former RIKEN Genomic Sciences Center, I began analyzing proteins using NMR spectroscopy in 1998. I am currently working on analyzing metabolites using NMR spectroscopy to explore metabolic activities.”
The term metabolism refers to cycles of chemical reactions carried out by cells in the body, and the resulting products are called metabolites. Metabolic activities in living organisms are constantly fluctuating, with the variations depending on the biological species. “In 2002, Dr Kurt Wüthrich of Switzerland received the Nobel Prize in Chemistry for his contributions to the development of NMR-based technology for protein structure analysis. At that time, I realized that the technology for protein structure analysis using NMR had reached its limit, so I needed to start something new. First, the genome, the ‘blueprint of life’, was decoded, and since then there have been constant advances in analyzing the structure of proteins produced on the basis of the genomic information. I thought of what could come next, and finally arrived at metabolic dynamics. I began analyzing metabolic dynamics using NMR spectroscopy around 2003. In those days, research of this kind was a great novelty around the world.”
In later years, research into metabolites rapidly attracted attention. In 2005, the Metabolomics Research Group (headed by director Kazuki Saito) was organized in the RIKEN Plant Science Center. Kikuchi joined the group as leader of the Advanced NMR Metabomics Unit, which was reorganized into his current team in 2010.
The products of metabolic processes are called metabolites, and the entire set of metabolites contained in a cell or individual organism is collectively referred to as its ‘metabolome’. The discipline of research into the metabolome is known as metabolomics.
However, Kikuchi’s team uses the term ‘metabomics’. “Doctor Jeremy Nicholson of the UK was the first researcher to undertake the analysis of metabolic dynamics using NMR spectroscopy. He called his study area ‘metabonomics’ to distinguish it from ‘metabolomics’. According to him, metabolomics refers to the ‘systematic study of metabolites’, with the emphasis placed on metabolites as they are, whereas metabonomics refers to the ‘systematic study of metabolism’, where urine and other biological products involving all metabolic activities in individual organisms are statistically analyzed to provide comprehensive insights into metabolic dynamics. What I want to do is to achieve a broad overview of metabolic fluctuations, which are complex, and this corresponds exactly to Nicholson’s concept of metabonomics. Since general awareness of both terms was low when I organized my team, I excluded both ‘-no-’ and ‘-lo-’ from the name of the team, but I did include this philosophical reasoning in the naming.
NMR enables comprehensive analysis of a wide variety of metabolites
There are many types of metabolites, including primary metabolites such as amino acids, sugars, lipids and nucleic acids, that are essential for the homeostasis, growth and reproduction of individual organisms, as well as secondary metabolites such as catechins and isoflavones, which are produced from primary metabolites. The properties of metabolites are similarly diverse. For example, some dissolve in water and others are insoluble, some are volatile, and some carry electric charges. “NMR spectroscopy allows us to analyze all metabolites, irrespective of whether they are soluble or insoluble, electrically charged or charge-free, or small molecules or macromolecules.”
An outline of the procedural flow for analyzing metabolites by NMR is shown in Figure 1. First, a ‘biofluid’ comprising a mixture of more than one metabolite of the organism to be examined, as it is or in the form of ‘squeezates’, is placed in a test tube, and the tube is set in an NMR instrument with an intense magnetic field. When the sample is irradiated with electromagnetic radiation, the atomic nuclei that make up the molecules of the metabolites absorb the radiation, exhibiting nuclear magnetic resonance. Electromagnetic radiation is emitted at a resonant frequency specific to each isotope. The resulting radiation is amplified and detected to determine the spectrum. A database of NMR spectra obtained by analyzing discrete metabolites allows identification of the metabolite corresponding to each spectrum. Peak intensity data show the quantity and motility of the metabolite. Metabolic dynamics can then be clarified by analyzing organisms for a variety of species and physiological states.
“The gold standard for successful analysis of metabolites is mass spectroscopy, but it requires purification and extraction of the single desired metabolite. My long-held desire was to observe something chaotic and complex in its original form, and I believed NMR could make this possible. In addition, not only the quantity of each metabolite, but also the composition ratio is important in evaluating metabolic dynamics. NMR enables us to determine the composition ratio of metabolites from the intensity ratio of the peaks that have been identified in the analysis of the metabolite mixture.”
Avoiding loss of samples and data
Nuclear magnetic resonance may be a revolutionary tool, but not all researchers are able to make the best use of its most important feature, which is enabling the analysis of metabolite mixtures. “When a sample containing insoluble metabolites is analyzed using conventional NMR, no useful data are obtained because of the low resolution. For this reason, people often dispose of the insoluble metabolites without performing analysis. However, metabolites disposed of in this way often carry useful information. I thought this was a waste. So in 2010, we developed a profiling method for the process of extracting metabolites from a mixture.” This method enables easy identification of which solvents to use in which order to achieve the most efficient extraction of metabolites. “It has become possible to obtain information on those metabolites that were previously disposed of as they are insoluble. Even for the insoluble metabolites that cannot be analyzed, high-resolution data can be obtained by employing high-resolution magic angle spinning, a sophisticated technique for taking measurements while rapidly spinning the sample at a specific angle.”
Kikuchi also developed the ‘comprehensive metabolic annotation’ method, which ensures that the data obtained can be used without any loss. The term ‘annotation’ refers to naming the spectrum obtained in relation to the assigned substance from a database. “Traditionally, spectral data has been disposed of if corresponding metabolites could not be found in any database. This is also a waste of useful information. With this new technique, we are able to statistically process the peak pattern data we have obtained, and select candidate metabolites with similar peak patterns from a database. Similarities in peak pattern are believed to suggest structural or functional similarities. Even though the substance may not be identical, the candidate may provide useful information if only its name includes an affix indicating that it is related to something.”
Previously, about 50 metabolites had been annotated on the basis of data obtained by analyzing metabolite mixtures using NMR spectroscopy. With the use of this new technique, the number has been increased to 211. “This is definitely a record-setting achievement,” says Kikuchi with pride.
Beyond metabolites toward macromolecular biomass
Kikuchi is now targeting macromolecular biomass. “In the case of plants, even when we apply the profiling method to the process of extracting metabolites, the residual biomass, which comprises a mixture of macromolecules such as cellulose and lignin, remains uncharacterized. Macromolecular biomass with molecular weights exceeding 1,000 is not usually referred to as metabolites. Biomass is always a mixture of macromolecules and insoluble components, so analysis is more difficult than with a metabolite mixture.” The term biomass refers to organic matter produced from carbon dioxide and water through photosynthesis in plants using solar energy, and thus it is a renewable resource. Kikuchi continues, “Today, in the twenty-first century, we are being called upon to reevaluate society’s dependence on finite resources such as petroleum and minerals, and to make a transition to a society oriented more toward the recycled use of biomass and other resources that can be renewed and that have a low cost per weight. Petroleum-derived products are not considered ‘organic’ in the consumer sense, whereas plant materials such as wood, bamboo and cotton have a natural beauty due to their nature as mixed substances. This creates a feeling of attachment. I want to establish a completely new analytical method for macromolecular biomass in order to make more efficient use of plant-derived biomasses.”
Kikuchi has recently been enthusiastically using the keyword ‘ecomics’. “It is a manifestation of my ambition to undertake research that will contribute to the resolution of environmental problems. In addition, removing the ‘no’ from ‘economics’ leaves ‘ecomics’. I am planning to initiate comprehensive research aimed at creating a society committed to recycling biological resources without overemphasizing economic activities.”
Mysterious relationships between probiotic and pathogenic bacteria
In January 2011, a study concerning the suppressive actions of the acetate produced by bifidobacteria on infection with the virulent O157 strain of Escherichia coli was widely covered in the media. The discovery was made by a joint research group mainly based around The University of Tokyo and the Laboratory for Epithelial Immunobiology (headed by Hiroshi Ohno) at the RIKEN Research Center for Allergy and Immunology, to which Kikuchi belongs. E. coli O157 is a pathogenic bacterium that causes food poisoning. Bifidobacteria, on the other hand, are probiotic bacteria that naturally inhabit the intestines, and are reportedly effective in maintaining good health. Although bifidobacteria were known to provide some defense against O157 infection, the underlying mechanisms were unknown.
“Bifidobacteria consume polysaccharides and produce acetic acid. In an experimental study using mice, we demonstrated that acetate protects the mucosal epithelium of the large intestine, enhancing the organ’s resistance to pathogens and hence suppressing O157 infection,” says Kikuchi (Fig. 2). Given this, taking vinegar would seem like an effective measure. However, vinegar is absorbed by the small intestine and does not reach the large intestine, and so is actually ineffective. Also, bifidobacteria can be obtained from yogurt and similar products, but their effect is low if taken as is. “A more effective approach is to ingest polysaccharides, or what are called dietary fibers, and bifidobacteria together. Our study clarified the relationship between probiotics and the polysaccharides that enhance their effects, and this represents a major achievement that is expected to find applications in maintaining health and preventing disease in humans.”
This study was followed by further investigations. “I wondered about the relationship between probiotic and pathogenic gut flora. You may believe that beneficial bacteria extirpate pathogens. However, we discovered an unexpected relationship.” Kikuchi analyzed metabolic dynamics while culturing O157 alone, bifidobacteria alone, and both O157 and the bifidobacteria together, in three test tubes set in an NMR instrument. One of the key features of NMR is the ability to analyze metabolic dynamics without killing the sample microbes, provided that they can be placed in test tubes.
The experiments revealed that the aspartic acid and serine produced by bifidobacteria were absorbed by O157, which in turn utilized these amino acids to produce succinate and acetate. The bifidobacteria and O157 were thus found to maintain a weak symbiotic relationship via the metabolites. “Since primary metabolites such as amino acids are produced in common by organisms, investigating how they function among different organisms has been difficult. In that study, making the best use of our own techniques for labeling metabolites with a stable carbon isotope, we found for the first time that amino acids play a key role in microbial symbiosis.”
Amino acids occur universally among organisms and they can be produced at low cost. “If unknown functions of primary metabolism can be clarified by making the best use of our new analytical method, we will be able to open new prospects in food science. Introducing changes in our sense of value and lifestyle in a way that will allow society to become more sustainable by distributing substances at lower manufacturing cost is similar in some way to efforts to build a sustainable society in which inexpensive biomass is recycled.”
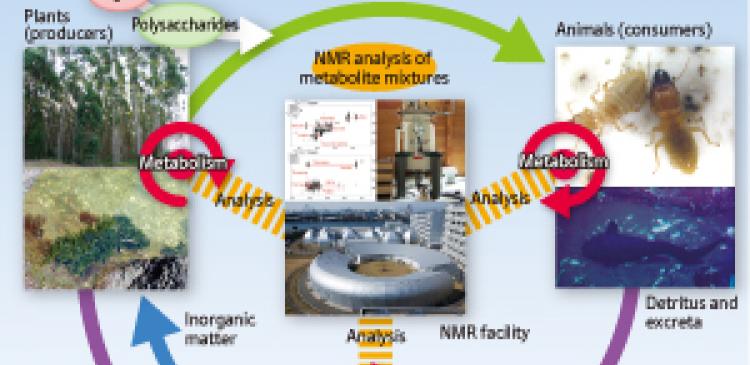
Kikuchi is looking at diverse targets, including not only higher plants such as rice and poplar, but also algae, complex microbial systems, insects, mice, fish, foods and even beverages (Fig. 3). “My approach may seem inconsistent, because I dabble in everything,” laughs Kikuchi, “but I want to achieve a broad view of metabolic dynamics in the biological world as a whole, as well as in individual organisms. Plants use photosynthesis to produce biomass from inorganic matter, animals ingest that biomass, and microorganisms decompose animal excreta and detritus into inorganic matter, which is returned to the soil. This process represents a profound world involving two aspects: ‘dynamism’, in which numerous substances are produced one after another by chemical reactions of small molecules; and ‘stillness’, in which macromolecular biomass remains accumulated as it is. In pursuing the laws of nature, I want to contemplate life through investigating the recycling of matter. For this, it is essential to analyze macromolecular biomass. I will give shape to this goal through my own efforts.”
Viewing cyclical changes in materials from the perspective of metabolic fluctuations. Individual organisms produce a wide variety of substances through the metabolic processes in their bodies. Plants use photosynthesis to produce organic matter from the inorganic, animals ingest that organic matter which they then move or disperse, and microorganisms decompose animal excreta and detritus back into the soil. In this way, materials move in a cycle. Kikuchi wants to reach a comprehensive view of biological life, not only from the viewpoint of metabolic dynamics in individual organisms, but also in terms of the recycling of materials among different organisms.