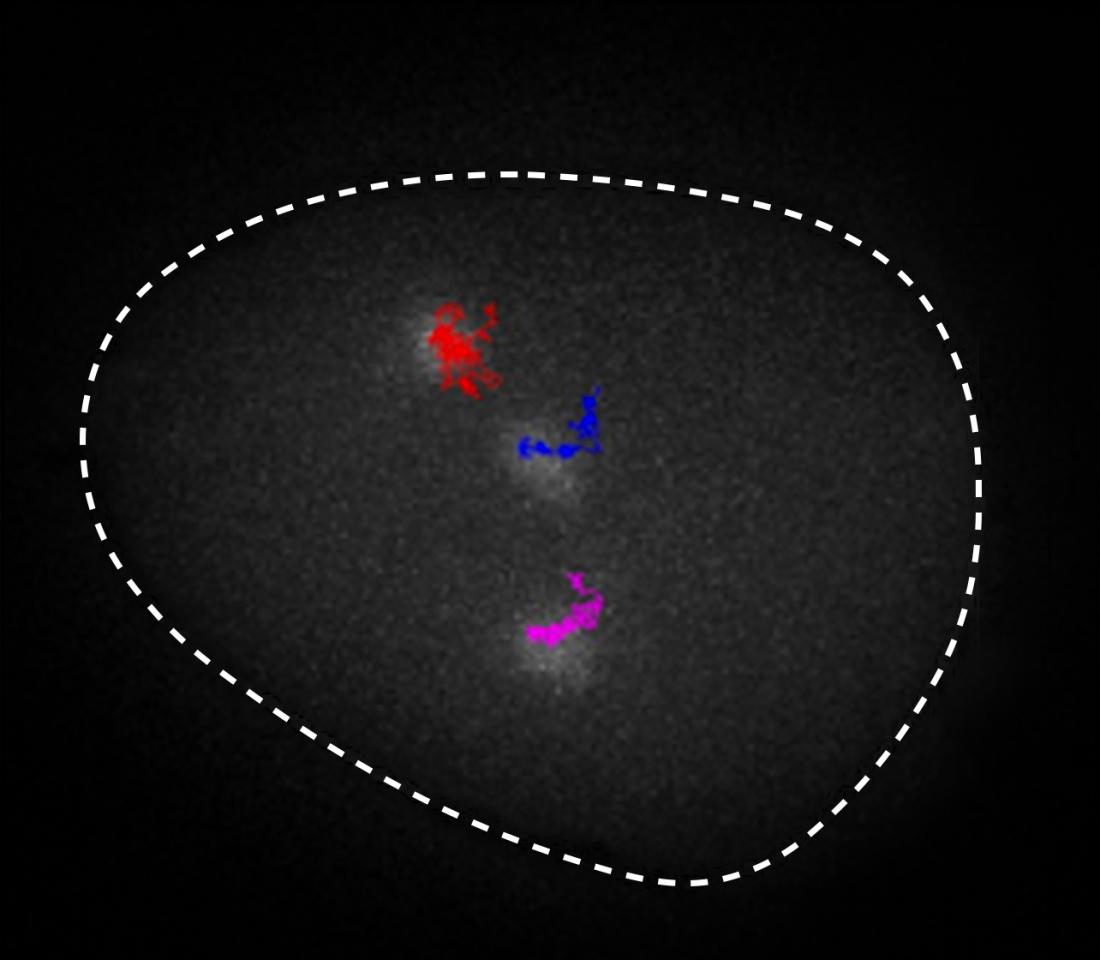
The new technique allows scientists to label specific points or regions of DNA in live cells and track their diffusing motions.
This story is featured in the Asia Research News 2021 magazine. Read in ISSUU (above) or full text and images below.
Researchers at the Ulsan National Institute of Science and Technology (UNIST) have developed a way to label specific points in DNA inside living cells with fluorescent probes, using the gene-editing technology CRISPR/ Cas9. The labelling system, described in the journal Genome Research, enables real-time tracking of how DNA is packaged inside the cell nucleus.
Each cell in the human body contains approximately two metres of DNA, efficiently wrapped around proteins called histones so it can fit inside the nucleus. The whole complex of DNA and histone proteins is called chromatin, and the physical structure and dynamics of chromatin heavily influence which genes are expressed.
“New methods for tracking the movements of chromatin in real time could help identify how changes in chromatin structure affect gene expression, aging and cancer,” says UNIST biomedical engineer Hajin Kim.
Existing techniques, such as Hi-C, have enabled scientists to infer the whole chromatin structure, but not to track specific sections along the DNA. The new technique allows scientists to label specific points or regions of DNA in live cells. Thus, it becomes possible to observe the structure of DNA regions (above) or to track their diffusing motions (lead photo at top of page).
Kim and his colleagues used the CRISPR/Cas9 gene editing technology to attach fluorescent probes to precise segments of DNA, called loci, inside living cells.
They were able to minimize background noise from the probes lighting up more than the targeted area to improve their ability to observe small sections of chromatin. They did this by combining a particular type of probe, called a tripartite superfolder green fluorescent protein, with another molecular system, called SunTag. This design amplifies the target signal and also allows certain components to be refreshed, extending the time the labelled loci glow.
Using the system, the researchers were able to see the location and motion of the target chromatin segment by looking through a fluorescence microscope. The team observed that DNA strands not only move passively inside the nucleus, as ink spreads in water, but also actively in certain time scales, confirming an earlier prediction, says Narendra Chaudhary, PhD candidate and first author of the study.
Now, researchers can use the system to target a wider range of loci throughout the genome and observe their movement inside the nucleus. It opens up new possibilities for studying genetic processes inside living cell nuclei, such as DNA replication, repair, recombination and transcription, through direct imaging.
Further information
Associate Professor Hajin Kim
[email protected]
Single Molecule Biophysics Laboratory
Ulsan National Institute of Science and Technology