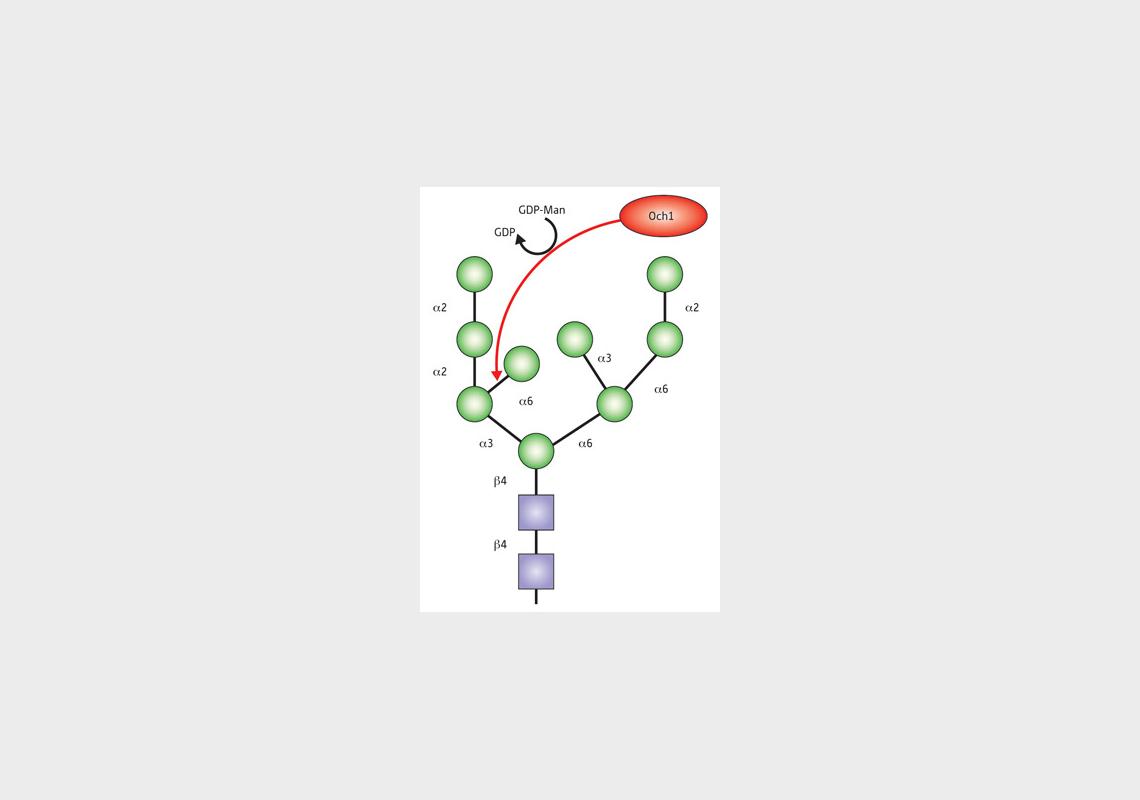
Schematic of a misfolded protein decorated with a diverse array of N-glycosylations prior to degradation in the ERAD pathway. Among these is a mannose carbohydrate introduced by the Golgi enzyme Och1, which might play a role in a novel late-stage protein ‘quality control’ system.
Many proteins undergo N-glycosylation, in which they are decorated with combinations of carbohydrates. These modifications not only contribute directly to normal protein function but also act as a flag for defective proteins, which get steered into a pathway known as endoplasmic reticulum-associated degradation (ERAD) with the assistance of enzymes that remove glycosylations to release free oligosaccharides (fOSs).
As an undergraduate, Tadashi Suzuki discovered the enzyme peptide:N-glycanase (PNGase) in the cytosol of mammalian cells; now, as a team leader at the RIKEN Advanced Science Institute, Wako, his group has uncovered valuable details about this enzyme’s critical contribution to the early stages of ERAD.
Suzuki and postdoctoral researcher Hiroto Hirayama recently turned to brewer’s yeast, a popular model organism, as a means to study an enigmatic PNGase-independent pathway for fOS production initially identified in mammalian cells. To achieve this, they developed an approach to selectively isolate these molecules, combining chemical labeling of oligosaccharides with a method for eliminating background contamination from β-1,6-glucans, a component of the yeast cell wall.
This strategy yielded a full library of yeast cytosolic fOSs—and some unexpected results. “To our surprise, we only detected PNGase-dependent fOSs under our experimental conditions,” says Suzuki. “This clearly indicates that mechanisms for generation of fOSs are quite distinct between mammals and yeast.”
To ensure that the full range of fOS diversity was represented, they performed their analysis in yeast lacking expression of the cytosol/vacuolar α-mannosidase (Ams1p), the only enzyme known to break down fOSs. “Very sophisticated and complicated glycan-recognition mechanisms for ERAD have been uncovered, but these conclusions have been drawn using a few model proteins,” says Suzuki. “On the other hand, we analyzed the whole population of fOSs, which means we can get information about glycan structures for all ERAD substrates.”
These data revealed that misfolded proteins can undergo diverse modifications prior to ERAD, including glycosylation by an enzyme within the Golgi apparatus, a cellular structure in which proteins typically undergo their final modifications. This suggests the existence of a previously unidentified screening mechanism at this late stage in protein synthesis that selectively redirects misfolded molecules to the ERAD system.
These and other findings suggest a great deal of hidden complexity remaining to be uncovered, and Suzuki’s team is now analyzing yeast strains with mutations in various proteins that help ‘read’ and interpret protein glycosylations. “Hopefully, through comparative fOS analysis for these strains, we can provide more precise mechanisms for the role of N-glycans in ERAD,” he says.
The corresponding author for this highlight is based at the Glycometabolome Team, RIKEN Advanced Science Institute