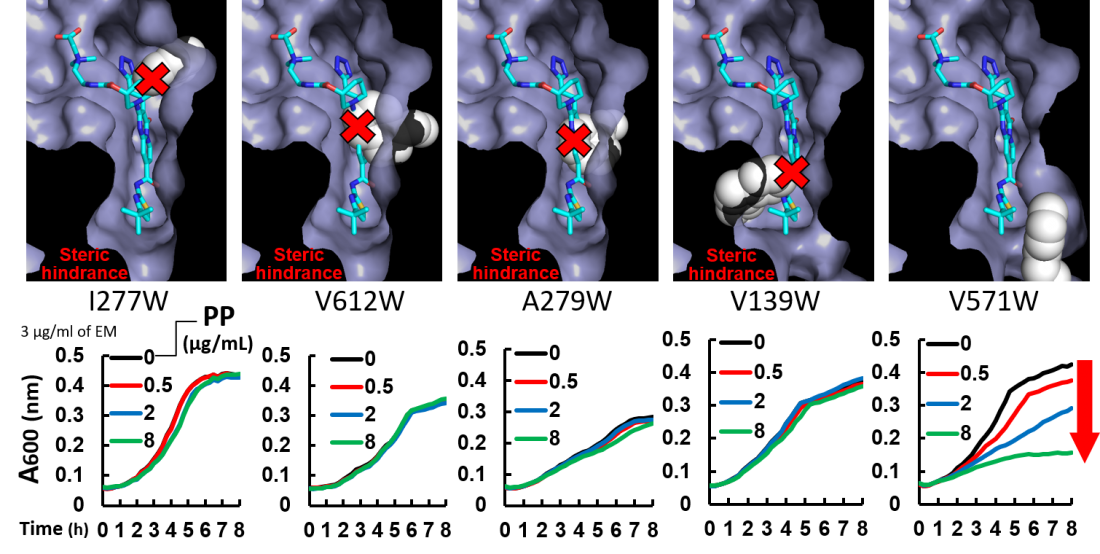
Evaluation of the putative structures of MexB mutants and the inhibitory effect of ABI-PP on the growth of E. coli expressing MexB mutants. (Top panels) Modeled closeup views of the inhibitor binding pit of ABI-PP-bound MexB mutants. The solvent accessible surface is depicted for each mutant, with the exception of mutated tryptophan side chains which are depicted as white, space-filling models. ABI-PP is shown as a stick model. (Bottom Panels) The inhibitory effect of ABI-PP on the growth of E. coli expressing each MexB mutant. Colored lines represent the degree of growth of each strain in the presence of the antibiotic erythromycin and one of several concentrations of PP (inhibitor ABI-PP). If the inhibitor can bind, increases in the inhibitor concentration result in corresponding degrees of inhibition of the pump, preventing antibiotic efflux and reducing bacterial growth (red arrows).
Researchers from SANKEN (The Institute of Scientific and Industrial Research), at Osaka University provide new insights into the relationship between the position of “bulky” amino acids on bacterial proteins and the ability of bacteria to resist antibiotics
Osaka, Japan – The medical profession is in the midst of losing an arms race. Bacterial antibiotic resistance doesn’t just threaten our ability to treat infection but our ability to carry out any treatment where infection is a risk. This includes a raft of life-saving surgeries ranging from coronary bypass operations to organ transplantation. In fact, the number of new antimicrobials being developed is declining each year. Understanding how bacteria resist the influence of antibiotics is essential to winning this arms race: it is time to make up ground.
In a study published this month in Antimicrobial Agents and Chemotherapy,researchers at Osaka University have produced new insights into the structure of a particular bacterial protein known as an efflux pump. This protein is involved in antibiotic resistance and its structure influences the ability of drugs to target it.
Many conventional antibiotics, such as penicillin or erythromycin, work by making their way into a bacterial cell. Once there, the antibiotic prevents the cell from working properly by interfering with its molecular machinery. From a bacterium’s perspective, this is where the efflux pump comes in handy; by pumping any compounds that are harmful to it (like antibiotics) out of the cell using an efflux pump, the bacterium can protect itself from them. Bacteria that have mutated to become particularly resistant to antibiotics often have many of these efflux pumps on their surface.
To counter this defense, researchers have produced a class of drugs that stop efflux pumps from working. These drugs, called efflux pump inhibitors, are effective at preventing the activity of some but not all types of efflux pump. Understanding how these drugs bind to efflux pumps is essential for understanding how they work, and so to producing new drugs.
Lead author Seiji Yamasaki says “We discovered the spatial characteristics of the inhibitor binding site in a bacterial efflux pump. We have succeeded in this analysis by examining the protein structure and by generating and analyzing a series of mutant pumps.”
The inhibitor-binding site of the wild-type MexB pump. (a) The crystal structure of the inhibitor ABI-PP bound to the MexB trimer. Three MexB monomers are shown in green, blue, and red, representing the access, binding, and extrusion monomer, respectively. ABI-PP is shown as a yellow space-filling model. (b) A close-up view of the inhibitor binding site. The substrate translocation pathway is shown as a solid gray surface. The proximal and distal binding pockets are indicated in green and blue circles, respectively. The inhibitor binding pit is shown as a red surface. The ABI-PP molecule is represented as a yellow stick model. (c) A detailed view of the inhibitor-binding site. Carbon atoms of ABI-PP are indicated in yellow while amino acid residues are indicated in green. The classification of these amino acids is shown on the right side of the panel.
By using genetic manipulation to change the position of a “bulky” amino acid called tryptophan, the researchers were able to show which positions were important for the ability of a specific inhibitor called ABI-PP to bind to, and prevent the action of, a specific efflux pump called MexB. Specifically, they found that bulky mutations to the top and the middle of the binding site were particularly effective at preventing ABI-PP binding.
Senior author Kunihiko Nishino says “Our results highlight that the overall spatial characteristics of the inhibitor binding site are more relevant to prevention of inhibition than single mutations. We hope that this work will inform the rational design of drugs that target efflux pumps, helping us to eliminate drug-resistant bacteria in the future.”
###
The article, “Spatial Characteristics of the Efflux Pump MexB Determine Inhibitor Binding,” was published in Antimicrobial Agents and Chemotherapy at DOI: https://journals.asm.org/doi/10.1128/aac.00672-22