Jellyfish have genetic adaptations that allow them to control their bodily salt concentration at the molecular level, allowing them to move vertically and horizontally to catch food without being affected by changing ocean salinity levels.
Featured in Asia Research News 2020 Magazine
The complete genome of Nemopilema nomurai, known as Nomura’s jellyfish, has been sequenced for the first time, providing clues to the genetic basis of the structural and biochemical traits that have allowed jellyfish to colonise our planet’s waters.
Nomura’s jellyfish, which can grow up to two metres in diameter, are found primarily around the coasts of China, Japan and South Korea. Since 2000, numbers have increased significantly, mainly due to global warming and a lack of natural predators.
Researchers at the Ulsan National Institute of Science and Technology (UNIST) and the Korea Institute of Ocean Science and Technology (KIOST) compared the genome of Nomura’s jellyfish with those of other animals in the Cnidaria phylum, a group that also includes hydra, sea anemones and corals.
“The cnidarians appear in a very important evolutionary position, as they share a common ancestor with the bilaterians, a major group of animals that includes humans, fish and insects, among others,” says UNIST geneticist Jong Bhak.
Between 500 and 700 million years ago, jellyfish developed novel physiological traits that allowed them to become one of the first actively swimming predators, rather than anchored in place or passively floating. To better understand these traits, the researchers studied genomic sequences and patterns of gene expression in the umbrella-shaped main body and tentacles of the Nomura’s jellyfish, as well as looking at the genes expressed during several jellyfish life stages. The results revealed genetic adaptations associated with swimming, including genes for muscle contraction and neuronal signalling, as well as an expansion of gene families coding for venom. These adaptations may have contributed to jellyfish mobility and active predation.
"Jellyfish have remarkable innovations that allow them to actively hunt in the water column."
One interesting finding was that jellyfish have genetic adaptations that allow them to control their bodily salt concentration at the molecular level, allowing them to move vertically and horizontally to catch food without being affected by changing ocean salinity levels.
“Jellyfish have remarkable innovations that allow them to actively hunt in the water column,” says Bhak. “Our study highlights the unique traits that have contributed to the success of these animals as early multi-cellular predators.”
The increasing prevalence and toxic venom of Nomura’s jellyfish means they pose a threat to fisheries and ocean ecosystems. Insights from this study could provide a basis to prevent jellyfish mass reproduction, as well as enhancing our understanding of cnidarian evolution.
Further information
Professor Jong Bhak | E-Mail: [email protected]
Korean Genomics Center
Ulsan National Institute of Science and Technology
Featured in Asia Research News 2020 Magazine
The many ways we can tell your research story. Find out more from our Content services page.
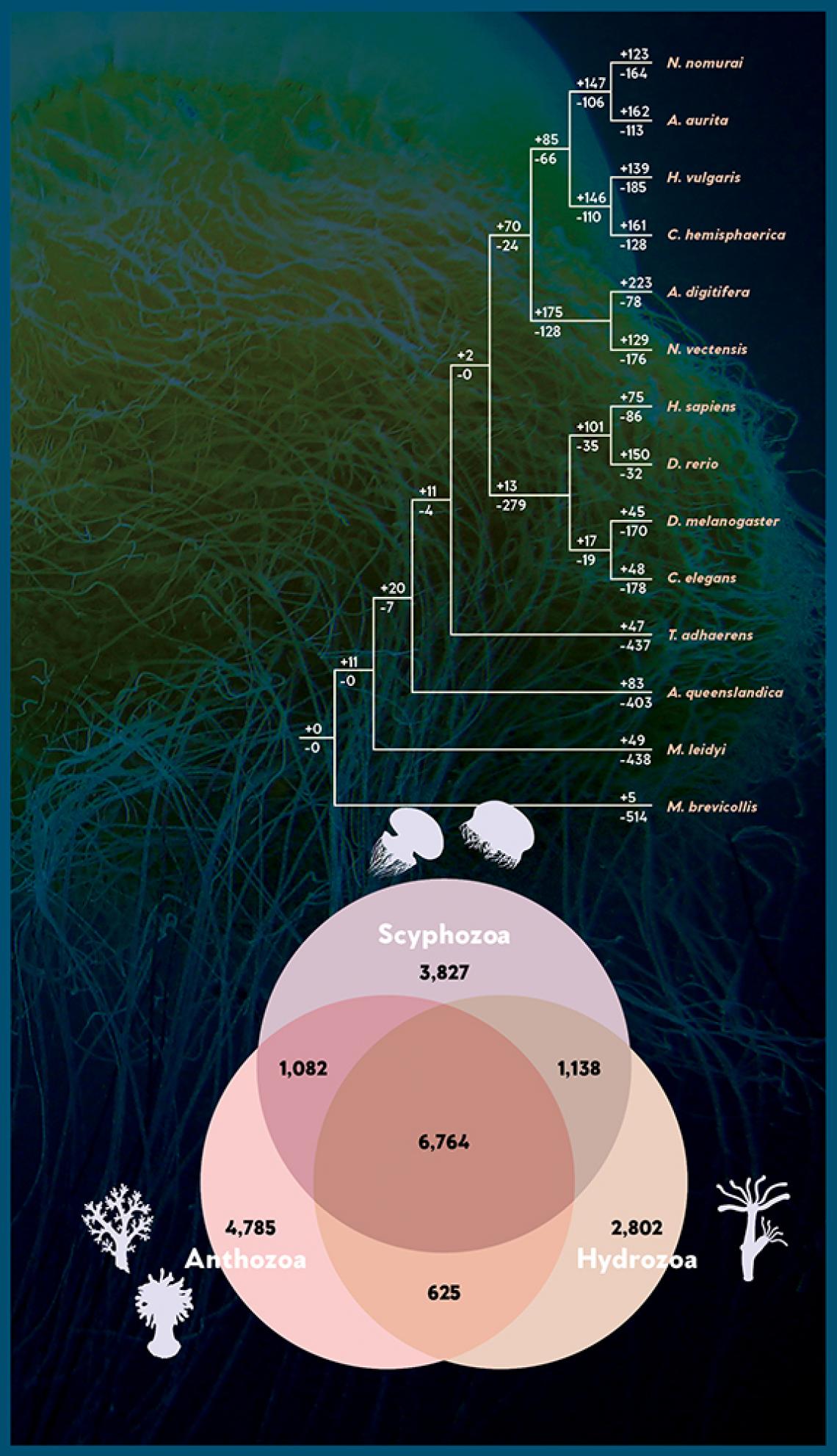
Gene family expansions and contractions in the Nemopilema genome. Numbers designate the number of gene families that have expanded (+) and contracted (−) after the split from the common ancestor.
Venn diagram shows the number of unique and shared gene families among three cnidarian classes: Scyphozoa, Hydrozoa and Anthozoa, which include jellyfish, corals and sea anemones.