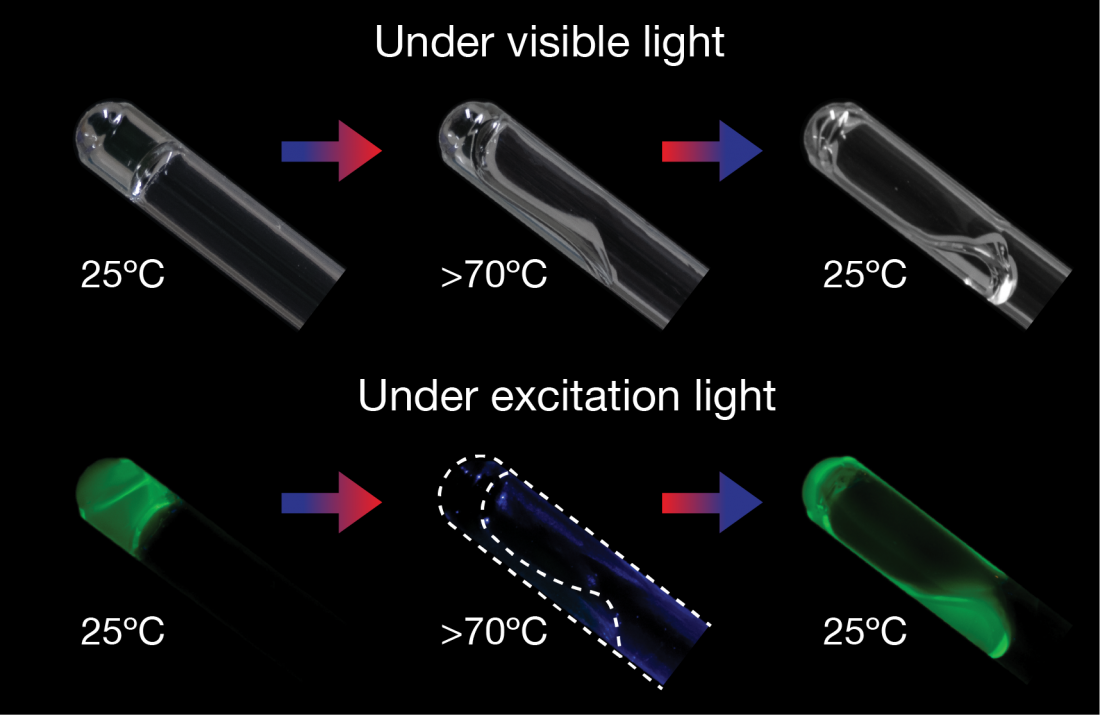
(Top) The star-polymer-DNA-gel (left) liquifies when its temperature is increased to more than 70˚C (center), and returns to a gel when the temperature drops back to 25˚C (right). (Bottom) Under UV light, the star-polymer-DNA-gel fluoresces green (left, right), but does not fluoresce when liquified (Photo: Xiang Li)
Scientists in Japan have made a tuneable, elastic and temperature-sensitive gel by using complementary DNA strands to connect star-shaped polymer molecules together. The gel, and the method used to develop it, could lead to advances in tissue regeneration, drug delivery and soft robotics. Xiang Li at Hokkaido University led the team of researchers who reported their findings in the journal Polymer Science.
Scientists have long been looking for better ways to develop gels that can be used in a variety of applications, including in the fields of medicine and engineering. Ideally, such gels need to be predictable in their behaviour, self-healing and durable enough for the rigorous jobs they are intended for.
“Gels are made by using bonds to link polymer molecules together,” explains Li. “When the bonds are connected, the material is more solid, and when they break in response to stress, the material turns to liquid.”
Owing to their high biocompatibility, water solubility and temperature sensitivity, DNA strands would be highly suitable for linking polymer molecules by taking advantage of their ability to form complementary bonds. However, scientists have so far found it difficult to use DNA links to develop homogeneous gels with on-demand elastic properties.
Looking to solve this problem, Li and his colleagues used software programs to simulate the formation of different DNA sequences and their complementary strands, and to determine how these double strands respond to changes in temperature. Their aim was to identify complementary DNA sequences that would only disconnect above 63°C in order to ensure a potential gel’s stability in the human body.
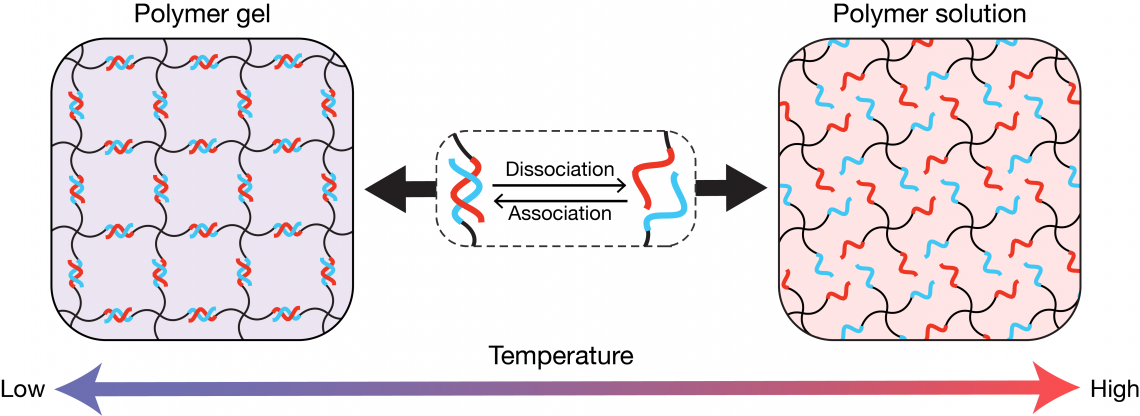
The star-polymer-DNA gel consists of complementary DNA strands (red and blue) linked to a molecule called polyethylene glycol (PEG, black). At lower temperatures, the DNA strands form complementary bonds leading to a gel; as the temperatures increase, the complementary bonds break and the gel liquifies. (Masashi Ohira, et al. Advanced Materials. January 16, 2022).
Based on the software simulations, they chose a pair of complementary DNA sequences to link four-armed molecules of polyethylene glycol (PEG). They prepared the gel by dissolving DNA strands and PEG separately in buffer solutions before mixing them in a test tube immersed in a hot water bath that was then cooled to ambient temperature. Finally, they conducted a series of experiments and analyses to evaluate the resulting gel’s properties.
The gel performed as predicted by the simulations, remaining elastic, self-repairing and solid until its melting temperature of 63°C over multiple testing cycles. The experiments also showed that the PEG molecules were homogeneously linked together by the DNA double strands and that liquid formation happened when the strands separated.
“Our findings suggest that we will be able to fabricate DNA gels with on-demand viscoelastic properties by making use of already available data on DNA thermodynamics and kinetics,” says Li. “The aim will be to improve the understanding and applications of this class of gel.”
Xiang Li, corresponding author, and Masashi Ohira, first author of this study (Photo: Xiang Li).