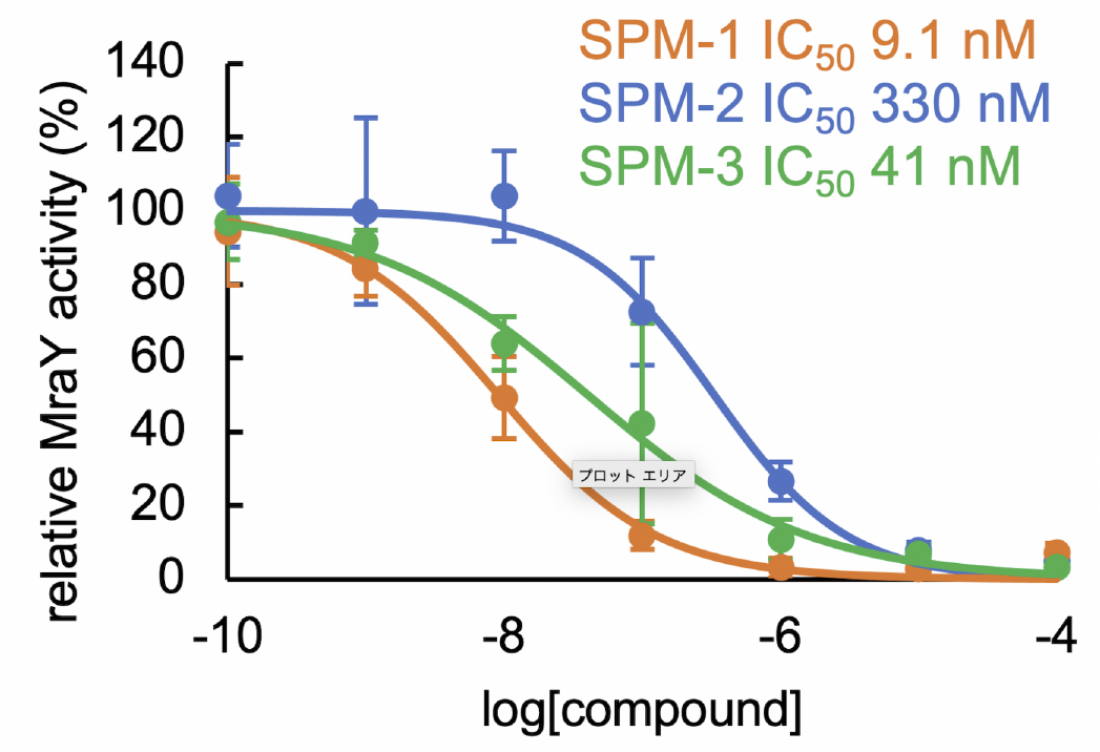
The sphaerimicin analogs (SPMs) inhibit the activity of MraY, and hence the replication of bacteria, with different degrees of effectiveness. The potency of the analog increases as the IC50 decreases (Takeshi Nakaya, et al. Nature Communications. December 20, 2022).
Antibiotics are vital drugs in the treatment of a number of bacterial diseases. However, due to continuing overuse and misuse, the number of bacteria strains that are resistant to multiple antibiotics is increasing, affecting millions of people worldwide. The development of new antibacterial compounds that target multiple drug resistant bacteria is also an active field of research so that this growing issue can be controlled.
A team led by Professor Satoshi Ichikawa at Hokkaido University has been working on the development of new antibacterials. Their most recent research, published in the journal Nature Communications, details the development of a highly effective antibacterial compound that is effective against the most common multidrug-resistant bacteria.
The team worked on a class of antibacterial compounds called sphaerimicins. These compounds block the function of a protein in the bacteria called MraY. MraY is essential for the replication of bacteria and plays a role in the synthesis of the bacterial cell wall; it is also not a target of currently available commercial antibiotics.
“Sphaerimicins are biological compounds, and have very complex structures,” explained Ichikawa, a corresponding author of the study. “We set out to design analogs to this molecule that would be easier to manufacture while also becoming more effective against MraY, thus increasing its antibacterial activity. The drug we designed was effective against methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus faecium (VRE), two of the more common multi-drug resistant bacteria.”
The team analysed structures of sphaerimicin A by molecular modelling assisted by calculation, and designed and synthesized two analogs of sphaerimicin, SPM1 and SPM2. These analogs were found to be effective against Gram positive bacteria.
They then determined the structure of SPM1 bound to MraY. By studying this structure and comparing it to that of related antibacterial agents, they determined how to further simplify the molecules. They were successful in developing a simpler analog, SPM3, whose activity was similar to SPM1.
In addition to their effectiveness against MRSA and VRE, the SPMs were also effective against Mycobacterium tuberculosis, the bacteria that causes tuberculosis—and which has multidrug-resistant strains.
“Our most significant contribution is the construction of the core skeleton of sphaerimicin, which can be used to develop more antibacterial agents that target MraY and hence multidrug resistant strains. Sphaerimicin is most promising as MraY is also present in Gram negative bacteria,” Ichikawa concluded. Future work will include optimisation of the currently developed SPM molecules, and the development of sphaerimicin-containing antibiotic combinations to target a wider range of bacteria.