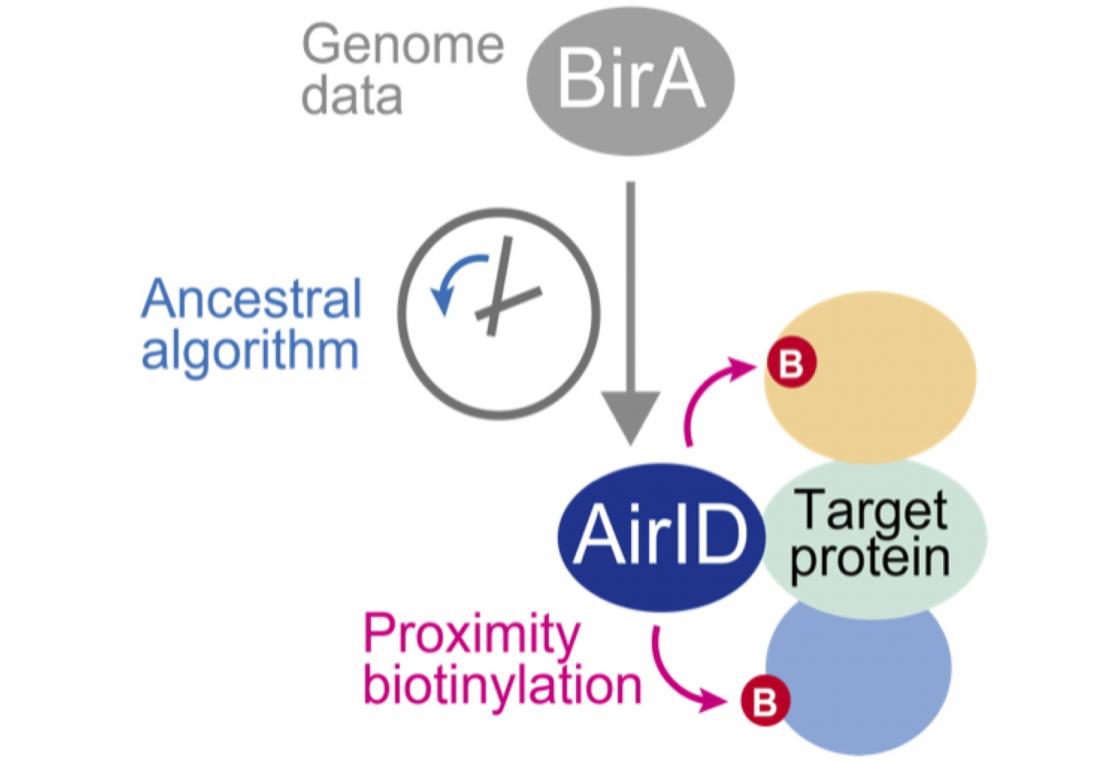
Fig. 1. Biotinylation of proteins interacting with a protein fused with AirID.
An ancestoral type enzyme AirID was designed from the sequence of biotinylation enzyme BirA of Escherichia coli by an ancestral enzyme reconstruction algorithm with metagenome data. Since AirID has high biotinylation activity and low cytotoxicity, it is possible to biotinylate a protein that interacts with the target protein by expressing it in cells as a fusion protein with the target protein. By identifying these biotinylated proteins using mass spectrometry analysis, it becomes possible to comprehensively identify proteins that interact with the target protein in cells.
Proteins control many physiological functions that are essential for living things in vivo. There are over 20,000 types of proteins in humans, and each protein plays a different role by interacting with other proteins. Proximity-dependent biotin identification (BioID) technology is a technique for finding these interacting partner proteins. BioID can be used by expressing the BioID-fusion protein and adding biotin. In cells expressing BioID-fusion bait protein, proteins with which the bait protein interacts are biotinylated and can be comprehensively analyzed using precipitation with streptavidin followed by mass spectrometry.
Three types of BioID enzymes (BioID, BioID2, TurboID) have been developed so far. However, BioID and BioID2 require a long reaction time and TurboID also biotinylates non-interacting proteins. Further improvement of biotinylation enzymes is an important goal for enhancing the convenience of proximity biotinylation in cells.
In this research, we designed ancestral biotinylation enzymes using an ancestral enzyme reconstruction algorithm and metagenome data. The new enzyme, AirID (ancestral BirA for proximity-dependent biotin identification) showed higher activity, higher specificity, and less toxicity than the previous three types. AirID was applied in the evaluation of protein interaction inhibitors, analyzing complexes formed by three or more types of proteins, and analyzing interactions through compounds called Molecular Glue. In addition, we have succeeded in the comprehensive identification of proteins that interact with a target protein by identifying biotinylated proteins using mass spectrometry analysis in cultured cells which express the target protein fused with AirID. These results indicate that AirID is a suitable enzyme for analyzing comprehensive protein–protein interactions in cells.